Innrain 80
6020 Innsbruck
Fax: +43 [Number]
Email: zlatko.trajanoski@i-med.ac.at
Website: http://icbi.at/
Research Branch (ÖSTAT Classification)
102004, 106005
Keywords
Bioinformatics, cancer immunology, colorectal cancer, and computational biology
Research Focus
The research of the Institute of Bioinformatics focuses on cancer immunology. Our aim is to decipher tumour-immune cell interaction by means of combined computational-experimental approaches. We specifically look at the question of how the immune system shapes the mutation spectrum of the tumour, both during progression and after treatment, in patients with colorectal cancer. We use patient-derived tumour models (organoids and tumour fragments), perturbation experiments and multimodal analysis, including next-generation sequencing, phosphoproteomics and imaging mass cytometry to facilitate precision immuno-oncology.
General Facts
The Institute of Bioinformatics was established in 2010 as the latest member of the Biocentre at the Medical University of Innsbruck and it is dedicated to supporting basic research and teaching in bioinformatics and computational biology. The Institute of Bioinformatics is well equipped for data processing and data management as well as computational analysis of a broad range of data types, including next-generation sequencing data, proteomics and metabolomics data and image analysis. It has a state-of-the-art computational infrastructure dedicated to bioinformatics analysis. A number of analytical pipelines, databases and software tools developed by the laboratory and others are installed and continuously maintained (see http://icbi.at).
Research
The long-term research goal of the Institute for Bioinformatics is to establish a mechanistic rationale for immunotherapy-based combination regimens and to develop a precision immuno-oncology platform for colorectal cancer. The concept integrates a living biobank of patient-derived tumour organoids with high-throughput and high-content data for testing drug combinations as well as machine learning in order to develop therapeutic recommendations for individual patients. We are investigating three immunomodulatory processes and their respective drug classes: 1) immuno-stimulatory effects of selected chemotherapeutics; 2) rewiring of signalling networks induced by targeted drugs and their interference with immunity; 3) metabolic reprogramming of T-cells to enhance anti-tumour immunity and its modulation by means of drugs.
For the first time, recent advancements in molecular profiling techniques, the development of organoid technology and novel computational methods, are providing the opportunity to tackle this challenging problem. Our systems-level dissection of cancer-immune cell interaction will facilitate the development of a recommendation algorithm to inform precision immuno-oncology in CRC and form the basis for an iterative platform to identify effective therapeutic regimens for individual patients.
As we have previously shown, the immune contexture – i.e. the density, composition, location and functional orientation of TILs – has a major prognostic value in CRC patients 1. Clinical observations supported by experimental evidence with mouse models indicate that chemotherapeutics as well as targeted drugs modulate the immune contexture and thus affect disease outcome (reviewed in 2). It has been provocatively hypothesised that the efficacy of most conventional and targeted anticancer drugs approved for use in humans is based not only on direct cytostatic/cytotoxic effects, but also on the reactivation of immune responses directed against the tumour 2. In accordance with this hypothesis, the goal of cancer therapy should be to restore immunological control of tumour growth. It is also notable that the mechanistic rationale behind the development of cancer immunotherapy differs from the rationale underlying conventional or targeted agents, i.e. reinstating immunological control of tumour growth rather than targeting tumour cells 2. Whilst this notion is gaining in popularity, examination of the complex interactions of tumours with the immune system has, until
recently, posed considerable challenges, owing to the lack of suitable cellular and animal systems and owing to the inadequacy of the available molecular tools for comprehensive immunological characterisation.
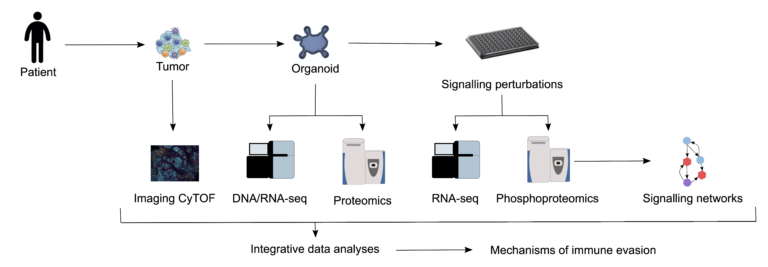
In the past, two key technologies were developed, which allow systematic investigation of the tumour-immune interface: 1) patient-derived tumour organoids that retain cell-cell and cell-matrix interactions, which more closely resemble those of the original tumour in comparison with cells grown in two dimensions on plastic dishes; 2) high-throughput and high-content technologies, including entire exome and RNA sequencing of bulk tissue, single-cell RNA sequencing, mass-spectrometry-based proteomics 3 and multiplexed staining of tissue sections. Moreover, considerable progress has been made in the development of novel computational tools for deep mining of the data. We use these tools (Fig. 1)
to investigate the impact of conventional and targeted drugs on T-cell activity, by means of archived samples, patient-derived organoids and systematic perturbation of the involved pathways and innovative computational tools. Our aim is to classify patients by using multidimensional profiling to analyse samples and to identify biomarkers of response to immunogenic chemotherapy and consequently patients who would benefit from combination therapy. Genomic, transcriptomic, phosphoproteomic and metabolomic characterisation of the organoids facilitates the generation of key quantitative data and will ultimately lead to the identification of effective drugs and drug combinations.
- Fridman WH, Pages F, Sautes-Fridman C, Galon J: The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer 2012, 12:298-306.
- Galluzzi L, Buque A, Kepp O, Zitvogel L, Kroemer G: Immunological Effects of Conventional Chemotherapy and Targeted Anticancer Agents. Cancer Cell 2015, 28:690-714.
- Aebersold R, Mann M: Mass-spectrometric exploration of proteome structure and function. Nature 2016, 537:347-355.
- Dienstmann R, Vermeulen L, Guinney J, Kopetz S, Tejpar S, Tabernero J: Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat Rev Cancer 2017, 17:268.
Pictures
Selected Publications
- Salcher, S.; Sturm, G.; Horvath, L.; Untergasser, G.; Kuempers, C.; Fotakis, G.; Panizzolo, E.; Martowicz, A.; Trebo, M.; Pall, G.; Gamerith, G.; Sykora, M.; Augustin, F.; Schmitz, K.; Finotello, F.; Rieder, D.; Perner, S.; Sopper, S.; Wolf, D.; Pircher, A.; Trajanoski, Z., High-resolution single-cell atlas reveals diversity and plasticity of tissue-resident neutrophils in non-small cell lung cancer. Cancer cell 2022, 40 (12), 1503-1520 e8.
- Rieder, D.; Fotakis, G.; Ausserhofer, M.; Rene, G.; Paster, W.; Trajanoski, Z.; Finotello, F., nextNEOpi: a comprehensive pipeline for computational neoantigen prediction. Bioinformatics 2021.
- Sturm, G.; Szabo, T.; Fotakis, G.; Haider, M.; Rieder, D.; Trajanoski, Z.; Finotello, F., Scirpy: a Scanpy extension for analyzing single-cell T-cell receptor-sequencing data. Bioinformatics 2020, 36 (18), 4817-4818.
- Yamazaki, T.; Kirchmair, A.; Sato, A.; Buque, A.; Rybstein, M.; Petroni, G.; Bloy, N.; Finotello, F.; Stafford, L.; Navarro Manzano, E.; Ayala de la Pena, F.; Garcia-Martinez, E.; Formenti, S. C.; Trajanoski, Z.; Galluzzi, L., Mitochondrial DNA drives abscopal responses to radiation that are inhibited by autophagy. Nature immunology 2020, 21 (10), 1160-1171.
- Monfort-Lanzas, P.; Gronauer, R.; Madersbacher, L.; Schatz, C.; Rieder, D.; Hackl, H., MIO: microRNA target analysis system for immuno-oncology. Bioinformatics 2022, 38 (14), 3665-3667.
Selection of Funding
FWF/DFG: Transregio 241 (DFG) Immune-Epithelial Communication in Inflammatory Bowel Diseases 2nd funding period (project partner: Z Trajanoski)
Horizon 2020, EIC 964955 Immune niches for cancer immunotherapy enhancement (project partner: Z Trajanoski)
Collaborations
Noel de Miranda, Leiden University, Leiden, The Netherlands
Florian Greten, Henner Farin, University of Frankfurt, Frankfurt, Germany