Innrain 80
6020 Innsbruck
Fax: +43 512 9003 73100
Email: Alexander.Huettenhofer@i-med.ac.at
Website: http://www.rnomics.at/
Research Branch (ÖSTAT Classification)
106002, 106014, 106023
Keywords
mitochondrial tRNAs, ncRNAs and disease, Non-coding RNAs, ribosome, RNA modifications, RNA sequencing (RNAseq), small nucleolar RNAs, splicing, and translation
Research Focus
In cells from all organisms two different types of RNA molecules are found: messenger RNAs, and so-called “non-protein-coding RNAs”. Many known ncRNAs are involved in the regulation of gene expression. Our research focuses on the identification of regulatory non-coding RNAs their functional characterization and their roles in human diseases. In addition to regulatory RNAs, the role of RNA modifications in mRNAs, tRNAs and rRNAs during protein synthesis are studied.
General Facts
The Institute of Genomics and RNomics is home to two research groups, which collaborate closely on various topics of RNA functions. The research interests range from the identification of regulatory ncRNAs and their functional characterization, as well their roles in human diseases by studying the roles of ncRNAs in splicing and various aspects of protein synthesis and RNA modifications. The common goal is a better understanding of the numerous functions and roles of different classes of RNAs. The Hüttenhofer group is focuses mainly on the characterization of the function of ncRNAs, which are involved in neurodegenerative diseases. Furthermore, recent projects imply the study of mitochondrial genetic elements found in the nuclear genome. The Erlacher group is focused on various aspects of protein synthesis, ranging from initiation to termination and its regulation by modified RNAs. Continuous third-party funding of the research groups (e.g. FWF stand-alone projects, participation in SFBs) enables the pursuit of various research projects. Alexander Hüttenhofer is head of the habilitation committee and part of the panel for good scientific practice. The Institute is participating in teaching in the curricula of molecular medicine, human medicine as well as in the PhD curriculum. We also participate in: “Die lange Nacht der Forschung” and”Open Lab days“, to introduce our research to the public and encourage young pupils to engage in the studies of Molecular Medicine.
Research
Project leader: Alexander Hüttenhofer
Roles of ncRNAs in human diseases
My group works on the identification and function of small non-coding RNAs (ncRNAs) in various model organisms which we have coined “Experimental RNomics“. By this approach, we are particularly interested in the involvement of ncRNAs in the etiology of human diseases. Small ncRNAs play important roles in the regulation of gene expression and have also been implicated in a number of diseases of the central nervous system (CNS).
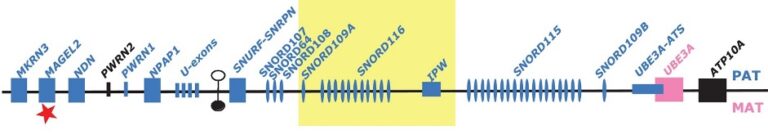
Prader-Willi Syndrome and Schaaf-Yang Syndrome: elucidation of molecular mechanisms (Figure 1)
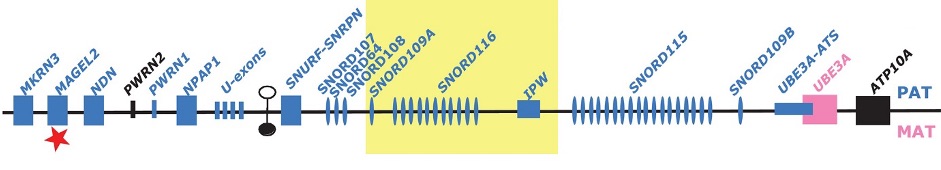
The genes of several brain-specific ncRNAs, which were first identified by my group, are located on chromosome 15. This region has been shown to be involved in the etiology to the Prader-Willi-Syndrome (PWS), a neuro-developmental disease. We demonstrated that these brain-specific ncRNAs are not expressed in PWS patients, pointing to their role in the PWS disease, which was later confirmed in PWS patients lacking these ncRNAs. We are currently investigating the role of these brain-specific ncRNAs in the etiology of PWS by various molecular biology approaches.
Chronic kidney disease:
Chronic kidney disease (CKD) is a progressive pathological condition marked by a gradual loss of kidney function. Exosomes, which are secreted from various cell types such as kidney cells, have emerged as a valuable repository of novel liquid biomarkers for human diseases. From CKD derived exosomes we have identified differentially expressed miRNAs, predominantly associated with the regulation of pro-inflammatory pathways as well as mitochondrial RNAs, highlighting the pivotal role of mitochondria in sensing renal inflammation.
NcRNAs and the SARS-Cov 2 virus infection
In collaboration with the Department for Hygiene and Microbiology (Prof. D. Wilflingseder and Prof. W. Posch), we are interested in investigating the small ncRNA transcriptome of lung epithelia cells infected with several different variants of the SARS-Cov2 virus (e.g. delta versus omicron variants). To that end, total RNA was prepared from non-infected and virus infected lung epithelial cells and an RNAseq analysis of the small ncRNA transcriptome of cellular ncRNAs or ncRNAs from the virus was analyzed.
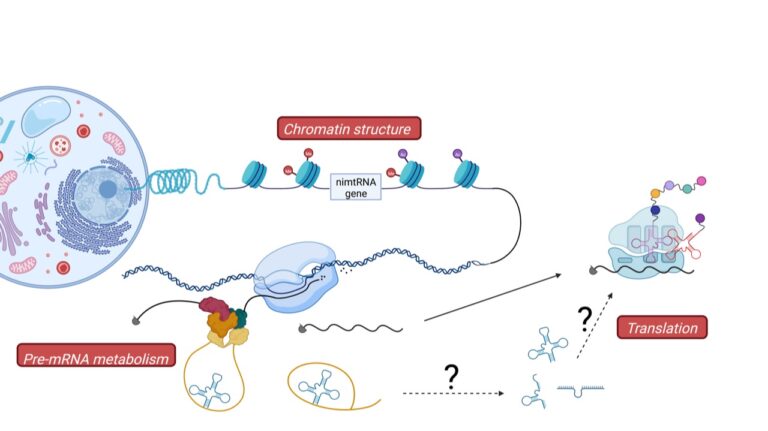
Novel functions of ncRNAs: evolutionary implications
During mammalian evolution, parts of the mitochondrial genome were integrated into the nuclear genome, including mitochondrial tRNAs (mtRNAs), also found within intronic regions of nuclear genes. We currently investigate the function of these nuclear intronic mitochondrial-derived tRNAs and their intergenic counterparts (Figure 2).
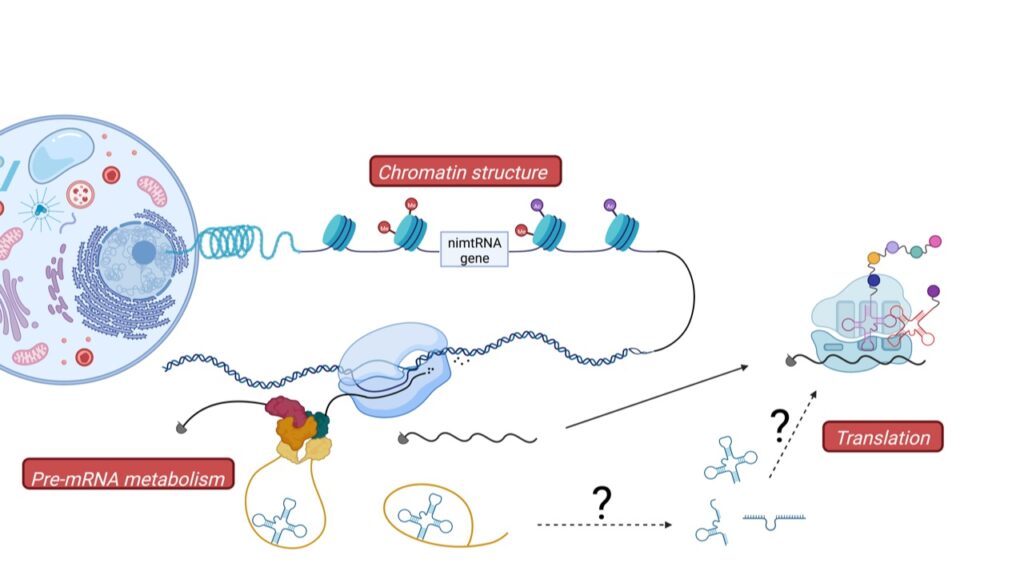
Indeed, we recently demonstrated that intronic mtRNAs are able to promote splicing of their host genes in analogy to splicing enhancer sequences. It thus appears that existing genes, e.g. mtRNA genes, were “exapted” during evolution into a novel function in the nucleus, namely in the regulation of pre-mRNA splicing.
Project leader Matthias Erlacher
RNA modifications as regulators of translation
More than 100 modified nucleotides have been identified in different classes of RNA. In many of them, the biological role is not well understood. By taking advantage of the developments in RNA chemistry, we generate synthetic tRNAs and mRNAs that harbor different natural and non-natural modifications. This approach allows the change of single chemical groups within the RNA of interest. These molecules are then systematically studied in different experimental settings, enabling a better understanding of essential processes such as translation initiation, elongation and termination.
DNA as template for ribosomal translation?
The central dogma of molecular biology describes the flow of the genetic information from DNA to RNA and ultimately to an amino acid sequence. One aim of this project is to extend the capability of the ribosome to also use single stranded DNA or other nucleotide derivatives as a template for protein synthesis. By applying an in vitro selection system using different rRNA libraries, we will select for different mutant ribosomes that will finally be able to shortcut the central dogma of biology. This will not only add a novel method to the toolbox of synthetic biology, but will also provide better insights into the decoding process and the “communication” within ribosome.
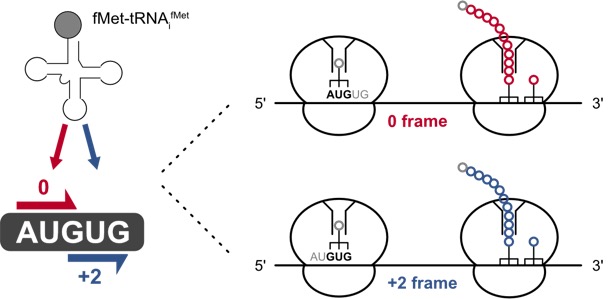
Ambiguous start sites: setting the right frame.
During initiation, the ribosome is tasked to efficiently recognize Open Reading Frames (ORFs) for the accurate and fast translation of mRNAs. The start codon recognition in bacteria, which is modulated by initiation factors, mRNA structure, the Shine Dalgarno (SD) sequence and the start codon itself is a critical step. Within the E. coli genome, we identified more than 50 annotated initiation sites harboring AUGUG or GUGUG sequence motifs that provide two canonical start codons, AUG and GUG, in immediate proximity. As these sites challenge the start codon recognition in E. coli, we study whether and how the ribosome is accurately guided to the designated ORF, providing the cell with sufficient initiation accuracy (Figure 3).
Pictures
Selected Publications
Maximilian P. Kohl, Maria Kompatscher, Nina Clementi, Lena Holl, Matthias D. Erlacher. Initiation at AUGUG and GUGUG sequences can lead to translation of overlapping reading frames in E. coli (2023). NUCLEIC ACIDS RESEARCH. 2023; 51(1):271-289;
Nir, Ronit; Hoernes, Thomas Philipp; Muramatsu, Hiromi; Faserl, Klaus; Kariko, Katalin; Erlacher, Matthias David; Sas-Chen, Aldema; Schwartz, Schraga: A systematic dissection of determinants and consequences of snoRNA-guided pseudouridylation of human mRNA.
NUCLEIC ACIDS RESEARCH. 2022; 50(9); 4900-4916.
Ranches, Glory; Zeidler, Maximilian; Kessler, Roman; Hoelzl, Martina; Hess, Michael W.; Vosper, Jonathan; Perco, Paul; Schramek, Herbert; Kummer, Kai K.; Kress, Michaela; Krogsdam, Anne; Rudnicki, Michael; Mayer, Gert; Huettenhofer, Alexander: Exosomal mitochondrial tRNAs and miRNAs as potential predictors of inflammation in renal proximal tubular epithelial cells.
MOLECULAR THERAPY-NUCLEIC ACIDS. 2022; 28(S); 794-813.
Krasheninina, Olga A.; Thaler, Julia; Erlacher, Matthias D.; Micura, Ronald: Amine-to-Azide Conversion on Native RNA via Metal-Free Diazotransfer Opens New Avenues for RNA Manipulations.
ANGEWANDTE CHEMIE-INTERNATIONAL EDITION. 2021; 60(13); 6970-6974.
Alisa O. Mikhaylina, Ekatarina Y Nikonova, Olga S. Kostareva, Wolfgang Piendl, Matthias D. Erlacher, Svetlana V. Tishchenko. Characterization of regulatory elements of L11 and L1 operons in thermophilic bacteria and archaea. BIOCHEMISTRY (Mosc). 2021; 86(4): 397-408. doi: 10.1134/s0006297921040027
Selection of Funding
FWF P34132; TAI411;
Collaborations
Peter Stadler, Leipzig University, Germany
Heiner Schaal, Heinrich Heine University Düsseldorf, Germany
Jürgen Brosius, University of Münster, Germany
Utz Fischer, University of Würzburg, Germany
Eric Westhof, Université de Strasbourg, France
Schraga Schwartz, Weizmann Institute of Science, Israel
Ronald Micura, University of Innsbruck, Austria
Christoph Kreutz, University of Innsbruck, Austria