Peter-Mayr-Straße 4b
6020 Innsbruck
Email: virologie@i-med.ac.at
Website: https://www.i-med.ac.at/virologie
Research Branch (ÖSTAT Classification)
303034, 301906, 301902, 302091, 301904, 301903
Keywords
cancer immunotherapy, clinical virology, immunogenic cell death, innate immunity, vaccines, virological diagnostic, Virology, and virotherapy
Research Focus
The major focus of the Institute of Virology is to develop novel biopharmaceuticals and vaccines. This research spans from elucidating the modes of action of novel therapeutics to the clinical translation and development in the spin-off biotech companies of the Institute.
Specific foci are:
>Virus-based oncolytic cancer vaccine strategies and their mode of action.
>Viral vector-based vaccines primarily against HIV and HPV.
>Antiviral immune response
General Facts
Structure: The Institute of Virology has two professors: Dorothee von Laer, the director, and Heribert Stoiber, the deputy director. In addition, there are five junior group leaders: Dr. Janine Kimpel, vector vaccines, Dr. Guido Wollmann, oncolytic viruses, and Dr. Wegene Borena, clinical virology, associated with D.v.Laer, Dr. Zoltan Banki,T-cell immunity, as well as Dr. Emmanuel Heilmann, viral evolution. The Christian Doppler Laboratory for Viral Immunotherapy of Cancer (Dr. G Wollmann) was established in 2017 within the institute.
Clinical routine: around 30% of the employees work in the serological and virological diagnostics group, which services the university hospital Innsbruck (LKI), the regional hospitals and the medical practices in Tirol. During the first wave of the SARS-CoV-2 pandemic, the diagnostics lab was at the forefront of high throughput PCR testing (> 1000 tests per day) in the state of Tyrol.
Collaborations: The division has developed an oncolytic viral cancer vaccine as well as complement enhanced therapeutic antibodies. To drive these two developments into clinical application, two companies were founded, ViraTherapeutics GmbH (founder D.v.Laer) and Lysovac (founder H. Stoiber), respectively. ViraTherapeutics has been acquired by Boehringer Ingelheim in September 2018.
The division is part of the European HIV Alliance (EHVA) funded by the European Union’s Horizon 2020 Research and Innovation Programme and collaborates with several international groups (s.8.) and in additions with groups and clinics in Innsbruck: Haematology and Oncology (Wolf), Cell Genetics (Baier, Kleiter), Bioinformatics (Trajanoski), Pathophysiology (Geley), ENT (Riechelmann, Schartinger), Urology (Culig, Horninger), Dermatology (Stoitzner) a.o.
Core facility: The division established and is now in charge (Dr. Janine Kimpel) of the BSL2 and BSL3 animal facilities of Innsbruck Medical University including an in-vivo imaging system (IVIS PerkinElmer).
Research
Covid 19: Epidemiology and antiviral Immunity
Janine Kimpel, Wegene Borena, Zoltan Banki
Since onset of the COVID-19 pandemic, we have established several assays to analyse antibody and T cell responses against SARS-CoV-2 and generated virus isolates for major SARS-CoV-2 variants (Riepler et al. 2020 Viruses (Basel), Borena et al. 2021 Viruses (Basel), Rössler et al. 2023 Lancet Microbe). These tools have been used for diagnostics as well as several epidemiologic studies to evaluate immunity, e.g. in the early hotspot Ischgl (Knabl et al. 2021 Commun Med (Lond), Borena et al. 2021 EBioMedicine, Pichler et al. 2021 J Infect Dis), in the district of Schwaz after the beta variant outbreak or after the ring vaccination campaign following this outbreak (Paetzold et al. 2022 Nat Commun, Winner et al. 2022 Euro Surveill, Willeit et al. 2022 Front Public Health, Harthaller et al. 2022 Viruses (Basel), Banki et al. 2022 Viruses (Basel), Seekircher et al. 2023 Lancet Microbe). Additionally, we conducted a clinical trial to analyse safety and immunogenicity of a heterologous COVID-19 vaccination combining one dose of a vector with a second mRNA dose, the HEVACC study (Banki et al. 2022 EBioMedicine, Rössler et al. 2022 Infect Dis, Loacker et al. 2023 Clin Chem Lab Med).
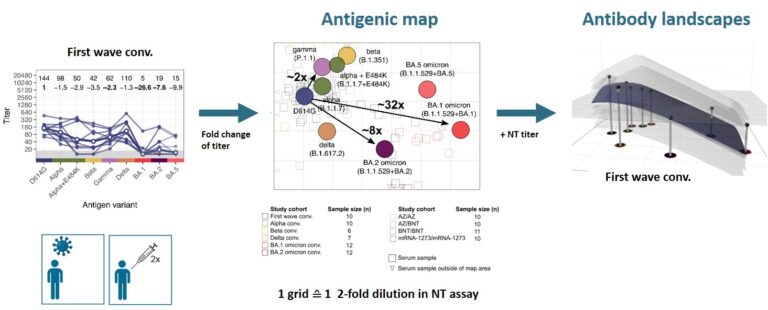
After the omicron variant emerged, we confirmed its strong immune escape indicating that omicron represents a distinct SARS-CoV-2 serotype (Rössler et al. 2022a N Engl J Med, Rössler et al. 2022b N Engl J Med). To better visualise antigenic relations of SARS-CoV-2 variants, we teamed up with Derek Smith from the University of Cambridge and performed antigenic cartography.
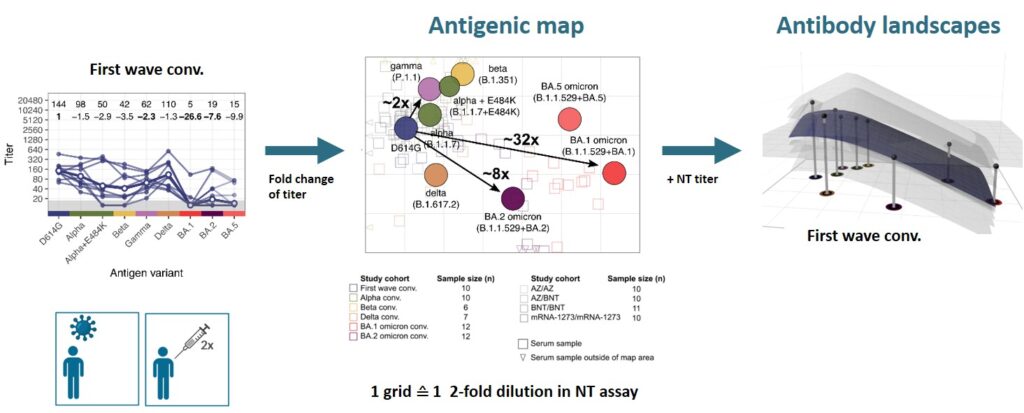
We found that cross-neutralising antibodies are induced after multiple exposures, especially with antigenically distant variants (Rössler et al. 2022 Nat Commun). Antigenic characterisation of emerging variants and neutralisation profiles after vaccination have important implications for development and selection of updated COVID-19 vaccines, which is why we were invited to present these data in WHO meetings discussing composition of updated COVID-19 vaccines.
Viral Vector Vaccines
Janine Kimpel
We have previously described the viral vector VSV-GP as promising candidate viral vector vaccine for infectious diseases such the human immunodeficiency virus HIV or the respiratory syncytial virus RSV. We continue to evaluate VSV-GP as HIV vaccine by using next-generation Envelope antigens and combining the vector in heterologous prime/boost regimens with other viral or non-viral vaccine platforms. Additionally, VSV-GP is also explored as a therapeutic vaccine against infections with the human papillomavirus (HPV). The idea here is to induce a T cell response against HPV proteins and thereby eliminate infected (pre)-malignant cells (Riepler et al. 2023 J Mol Biol).
Oncolytic Viruses
Guido Wollmann
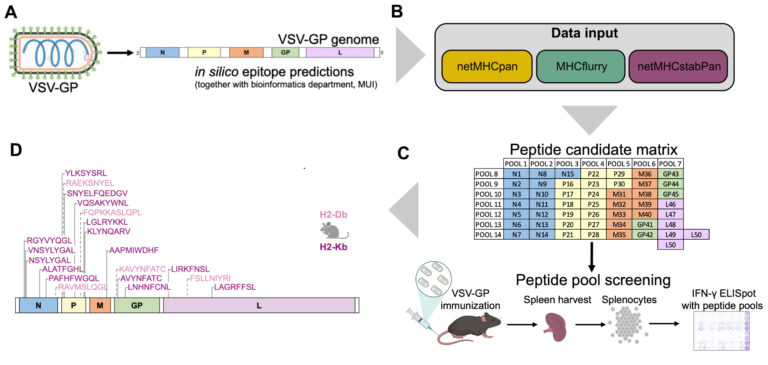
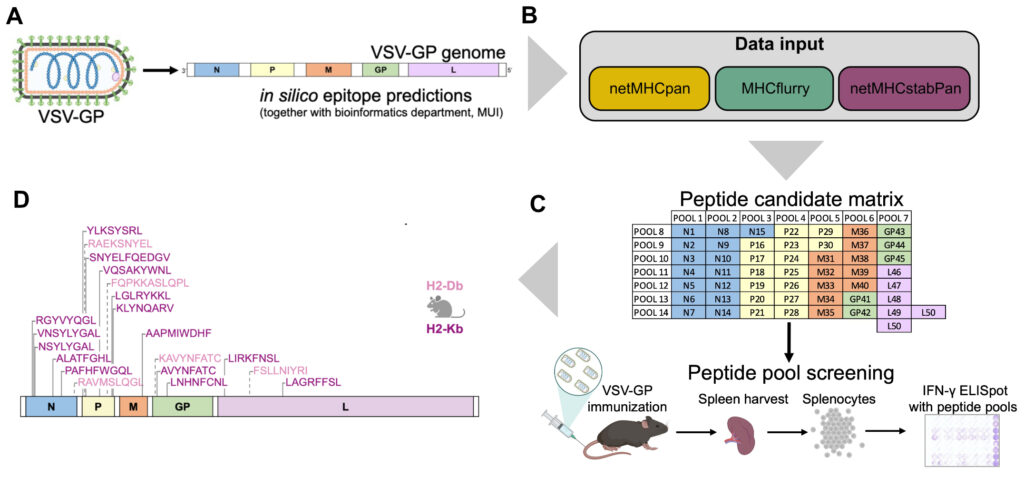
The oncolytic virus (OV) VSV-GP that was developed in our institute is now entering clinical trial testing. We are currently establishing immuno-monitoring tools based on single cell sequencing and viral epitope predictions that can be applied in clinical trials to better characterise the antiviral immune response. Further, we develop next generation OVs with altered biology with a specific focus on inducing immunogenic cell death (ICD), such as necroptosis and ferroptosis. In the optimal setting, ICD holds the potential to induce an antitumor immune response even when tumour-antigens are not known, leading to in-situ cancer vaccination. Similarly, we are also exploring whether the addition of ICD can enhance designed cancer vaccines with known antigens.
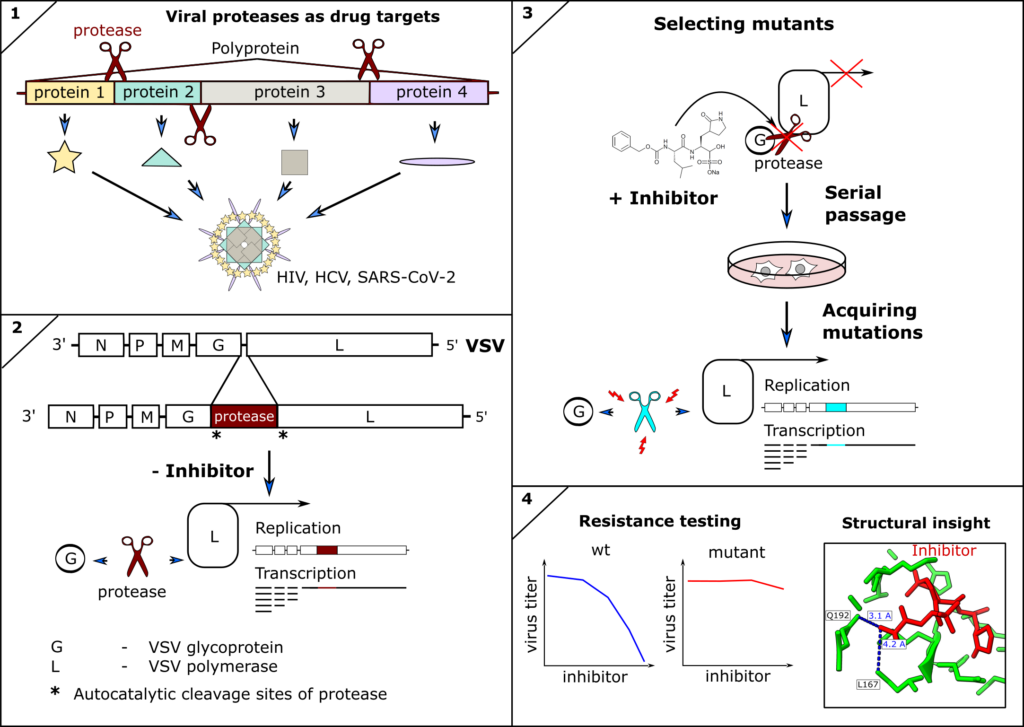
Viral Proteases as therapeutic targets
Emmanuel Heilmann
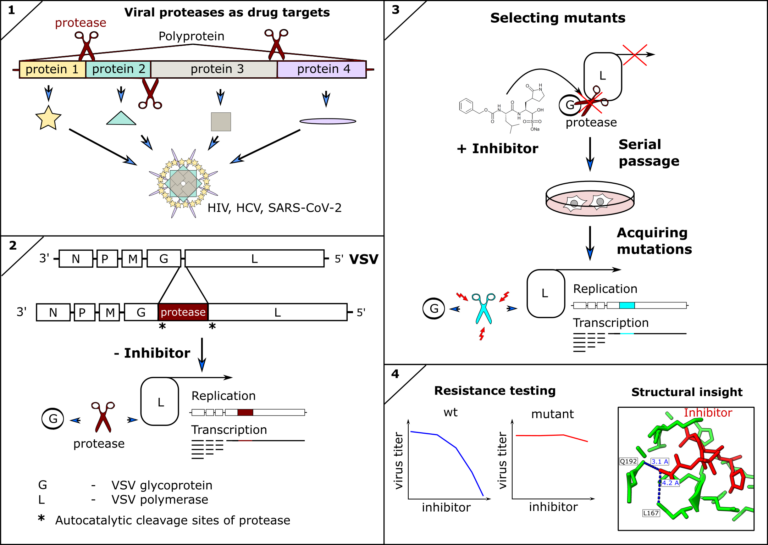
Inhibitors of viral proteases have proven to be highly potent antivirals against human immunodeficiency virus (HIV), hepatitis-C virus (HCV), and recently SARS-CoV-2. However, antivirals and indeed any antimicrobial drug can lose efficacy if escape mutants of the drug target occur and are fixed within the pathogen population. Therefore, surveillance and resistance profiling can guide therapeutic decisions, such as combining treatments to restore efficacy. Furthermore, structural information of mutant proteins informs targeted chemical modification of drugs to circumvent resistances. Ideally, resistance profiles are available before mutations become clinically relevant. We therefore engineer(ed) novel tools to predict and test mutations of the SARS-CoV-2 and other coronavirus proteases against clinically approved inhibitors. We described the first antiviral resistance mutations against Paxlovid as predicted by our methods (DOI: 10.1126/scitranslmed.abq7360) as well as already circulating SARS-CoV-2 antiviral resistant mutants (DOI: 10.1126/sciadv.ade8778).
Virus complement interaction
Heribert Stoiber
As a member of a nation-wide consortium, we evaluated the performance of the antigen-based anterior nasal screening program implemented in all Austrian schools to detect SARS-CoV-2 infections. A nationwide antigen-based screening combined data obtained in March 2021 from 5,370 schools (Grade 1-8) with an RT-qPCR-based prospective cohort study comprising a representative sample of 244 schools. This monitoring study in Austrian schools revealed SARS-CoV-2 infection in of participants and identified associations of regional community incidence and social deprivation with higher prevalence.
In addition, the interaction of Complement proteins with Zikavirus (ZIKV) was investigated. By virus-capture assay and Western blots, the complement regulator protein CD55 was identified to be incorporated into the viral envelope. Blocking of CD55 by neutralising Abs significantly increased the sensitivity to human complement. These data indicate that the incorporation of CD55 from human cells contributes to the stability of ZIKV against complement-mediated virolysis.
Pictures
Selected Publications
Janine Kimpel
- Rössler A, Knabl L, Raschbichler LM, Peer E, von Laer D, Borena W, Kimpel J. Reduced sensitivity of antibody tests after omicron infection. Lancet Microbe. 2023 Jan;4(1):e10-e11. doi: 10.1016/S2666-5247(22)00222-1. Epub 2022 Sep 19.
- Rössler A*, Netzl A*, Knabl L, Schäfer H, Wilks SH, Bante D, Falkensammer B, Borena W, von Laer D, Smith DJ*, Kimpel J*.2 and BA.5 omicron differ immunologically from both BA.1 omicron and pre-omicron variants. Nat Commun. 2022 Dec 13;13(1):7701. doi: 10.1038/s41467-022-35312-3.
- Bánki Z*, Mateus J*, Rössler A*, Schäfer H, Bante D, Riepler L, Grifoni A, Sette A, Simon V, Falkensammer B, Ulmer H, Neurauter B, Borena W; HEVACC Study Group; Krammer F*, von Laer D*, Weiskopf D*, Kimpel J*. Heterologous ChAdOx1/BNT162b2 vaccination induces stronger immune response than homologous ChAdOx1 vaccination: The pragmatic, multi-center, three-arm, partially randomized HEVACC trial. 2022 Jun;80:104073. doi: 10.1016/j.ebiom.2022.104073.
- Rössler A, Knabl L, von Laer D, Kimpel J. Neutralization Profile after Recovery from SARS-CoV-2 Omicron Infection. N Engl J Med. 2022 May 5;386(18):1764-1766. doi: 10.1056/NEJMc2201607. Epub 2022 Mar 23.
- Rössler A, Riepler L, Bante D, von Laer D, Kimpel J. SARS-CoV-2 Omicron Variant Neutralization in Serum from Vaccinated and Convalescent Persons. N Engl J Med. 2022 Feb 17;386(7):698-700. doi: 10.1056/NEJMc2119236. Epub 2022 Jan 12. PMID: 35021005 Free PMC article.
- Heilmann E, Costacurta F, Moghadasi SA, Ye C, Pavan M, Bassani D, Volland A, Ascher C, Weiss AKH, Bante D, Harris RS, Moro S, Rupp B, Martinez-Sobrido L, von Laer D. SARS-CoV-2 3CLpro mutations selected in a VSV-based system confer resistance to nirmatrelvir, ensitrelvir, and GC376. Sci Transl Med. 2023 Jan 11;15(678):eabq7360. doi: 10.1126/scitranslmed.abq7360. Epub 2023 Jan 11.
- Moghadasi SA, Heilmann E, Khalil AM, Nnabuife C, Kearns FL, Ye C, Moraes SN, Costacurta F, Esler MA, Aihara H, von Laer D, Martinez-Sobrido L, Palzkill T, Amaro RE, Harris RS. Transmissible SARS-CoV-2 variants with resistance to clinical protease inhibitors. Sci Adv. 2023 Mar 29;9(13):eade8778. doi: 10.1126/sciadv.ade8778. Epub 2023 Mar 29.
- Volland A, Lohmüller M, Heilmann E, Kimpel J, Herzog S, von Laer D. Heparan sulfate proteoglycans serve as alternative receptors for low affinity LCMV variants. PLoS Pathog. 2021 Oct 14;17(10):e1009996. doi: 10.1371/journal.ppat.1009996. eCollection 2021 Oct.
Guido Wollmann
- Vijver SV, Danklmaier S, Pipperger L, Gronauer R, Floriani G, Hackl H, Das K, Wollmann G. Prediction and validation of murine MHC class I epitopes of the recombinant virus VSV-GP. Front Immunol. 2023 Jan 19;13:1100730. doi: 10.3389/fimmu.2022.1100730.
- Boch C, Reschke M, Igney F, Maier P, Müller P, Danklmaier S, Das K, Hofer T, Wollmann G, Rist W. Evaluating Antibody Pharmacokinetics as Prerequisite for Determining True Efficacy as Shown by Dual Targeting of PD-1 and CD96. Biomedicines. 2022 Sep 1;10(9):2146. doi: 10.3390/biomedicines10092146.
- Hofer T, Rossi M, Carboni S, Di Berardino Besson W, von Laer D, Wollmann G, Derouazi M, Santiago-Raber ML.. 2021. Heterologous Prime-Boost Vaccination with a Peptide-Based Vaccine and Viral Vector Reshapes Dendritic Cell, CD4+ and CD8+ T Cell Phenotypes to Improve the Antitumor Therapeutic Effect. Cancers 13:6107
- Das K, Belnoue E, Rossi M, Hofer T, Danklmaier S, Nolden T, Schreiber LM, Angerer K, Kimpel J, Hoegler S, Spiesschaert B, Kenner L, von Laer D, Elbers K, Derouazi M, Wollmann G. A modular self-adjuvanting cancer vaccine combined with an oncolytic vaccine induces potent antitumor immunity. Nature Communications 2021, 12:5195
- Lechner M, Schartinger VH, Steele CD, Nei WL, Ooft ML, Schreiber LM, Pipinikas CP, Chung GT, Chan YY, Wu F, To KF, Tsang CM, Pearce W, Morelli D, Philpott M, Masterson L, Nibhani R, Wells G, Bell CG, Koller J, Delecluse S, Yip YL, Liu J, Forde CT, Forster MD, Jay A, Dudás J, Krapp A, Wan S, Uprimny C, Sprung S, Haybaeck J, Fenton TR, Chester K, Thirlwell C, Royle G, Marafioti T, Gupta R, Indrasari SR, Herdini C, Slim MAM, Indrawati I, Sutton L, Fles R, Tan B, Yeong J, Jain A, Han S, Wang H, Loke KSH, He W, Xu R, Jin H, Cheng Z, Howard D, Hwang PH, Le QT, Tay JK, West RB, Tsao SW, Meyer T, Riechelmann H, Oppermann U, Delecluse HJ, Willems SM, Chua MLK, Busson P, Lo KW, Wollmann G, Pillay N, Vanhaesebroeck B, Lund VJ. Somatostatin receptor 2 expression in nasopharyngeal cancer is induced by Epstein Barr virus infection: impact on prognosis, imaging and therapy. Nature Communications, 2021, 12:117
Wegene Borena
- Waser M, Heiss R, Borena W. Factors affecting children’s HPV vaccination in Austria: Evidence from a parent survey. Hum Vaccin Immunother. 2022 Nov 30;18(6):2126251. doi: 10.1080/21645515.2022.2126251. Epub 2022 Oct 17.
- Borena W, Banki Z, Bates K, Winner H, Riepler L, Rössler A, Pipperger L, Theurl I, Falkensammer B, Ulmer H, Walser A, Pichler D, Baumgartner M, Schönherr S, Forer L, Knabl L, Würzner R, von Laer D, Paetzold J, Kimpel Persistence of immunity to SARS-CoV-2 over time in the ski resort Ischgl. EBioMedicine. 2021 Aug;70:103534. doi: 10.1016/j.ebiom.2021.103534. Epub 2021 Aug 12.
- Harthaller T, Borena W, Bante D, Schäfer H, Strallhofer O, Zöggeler T, Hochmuth E, Hoch L, Rössler A, von Laer D, Kimpel J, Falkensammer High Prevalence of Undocumented SARS-CoV-2 Infections in the Pediatric Population of the Tyrolean District of Schwaz. Viruses. 2022 Oct 19;14(10):2294. doi: 0.3390/v14102294.
- Willeit P, Kimpel J, Winner H, Harthaller T, Schäfer H, Bante D, Falkensammer B, Rössler A, Riepler L, Ower C, Sacher M, von Laer D, Borena W. Seroprevalence of SARS-CoV-2 infection in the Tyrolean district of Schwaz at the time of the rapid mass vaccination in March 2021 following B.1.351-variant outbreak. Front Public Health. 2022 Sep 9;10:989337. doi: 10.3389/fpubh.2022.989337. eCollection 2022.
- Borena W, Kimpel J, Gierer M, Rössler A, Riepler L, Oehler S, von Laer D, Miholits Characterization of Immune Responses to SARS-CoV-2 and Other Human Pathogenic Coronaviruses Using a Multiplex Bead-Based Immunoassay. Vaccines (Basel). 2021 Jun 7;9(6):611. doi: 10.3390/vaccines9060611.
Zoltan Banki
- Bánki Z, Seekircher L, Falkensammer B, Bante D, Schäfer H, Harthaller T, Kimpel J, Willeit P, von Laer D, Borena Six-Month Follow-Up of Immune Responses after a Rapid Mass Vaccination against SARS-CoV-2 with BNT162b2 in the District of Schwaz/Austria. Viruses 2022 Jul 27;14(8):1642. doi: 10.3390/v14081642.
- Bánki Z, Mateus J, Rössler A, Schäfer H, Bante D, Riepler L, Grifoni A, Sette A, Simon V, Falkensammer B, Ulmer H, Neurauter B, Borena W, HEVACC Study Group, Krammer F, von Laer D, Weiskopf D, Kimpel Heterologous ChAdOx1/BNT162b2 vaccination induces stronger immune response than homologous ChAdOx1 vaccination: The pragmatic, multi-center, three-arm, partially randomized HEVACC trial. EBioMedicine. 2022 Jun;80:104073. doi: 10.1016/j.ebiom.2022.104073. Epub 2022 May 23.
- Borena W, Bánki Z, Bates K, Winner H, Riepler L, Rössler A, Pipperger L, Theurl I, Falkensammer B, Ulmer H, Walser A, Pichler D, Baumgartner M, Schönherr S, Forer L, Knabl L, Würzner R, von Laer D, Paetzold J, Kimpel J. Persistence of immunity to SARS-CoV-2 over time in the ski resort Ischgl. EBioMedicine. 2021 Aug;70:103534. doi: 10.1016/j.ebiom.2021.103534. Epub 2021 Aug 12.
- Pipperger L, Riepler L, Kimpel J, Siller A, Stoitzner P, Bánki Z, von Laer D. Differential infection of murine and human dendritic cell subsets by oncolytic vesicular stomatitis virus variants. Oncoimmunology. 2021 Aug 31;10(1):1959140. doi: 10.1080/2162402X.2021.1959140. eCollection
- Lackner M, Rössler A, Volland A, Stadtmüller MN, Müllauer B, Banki Z, Ströhle J, Luttick A, Fenner J, Sarg B, Kremser L, Tone P, Stoiber H, von Laer D, Wolff T, Schwarz C, Nagl M. N-chlorotaurine is highly active against respiratory viruses including SARS-CoV-2 (COVID-19) in vitro. Emerg Microbes Infect. 2022 Dec;11(1):1293-1307. doi: 10.1080/22221751.2022.2065932.
Dorothee von Laer
- Heilmann E, Costacurta F, Moghadasi SA, Ye C, Pavan M, Bassani D, Volland A, Ascher C, Weiss AKH, Bante D, Harris RS, Moro S, Rupp B, Martinez-Sobrido L, von Laer D. SARS-CoV-2 3CLpro mutations selected in a VSV-based system confer resistance to nirmatrelvir, ensitrelvir, and GC376. Sci Transl Med. 2023 Jan 11;15(678):eabq7360. doi: 10.1126/scitranslmed.abq7360. Epub 2023 Jan 11.
- Borena W, Bánki Z, Bates K, Winner H, Riepler L, Rössler A, Pipperger L, Theurl I, Falkensammer B, Ulmer H, Walser A, Pichler D, Baumgartner M, Schönherr S, Forer L, Knabl L, Würzner R, von Laer D, Paetzold J, Kimpel J. Persistence of immunity to SARS-CoV-2 over time in the ski resort Ischgl. EBioMedicine. 2021 Aug;70:103534. doi: 10.1016/j.ebiom.2021.103534. Epub 2021 Aug 12.
- Volland A, Lohmüller M, Heilmann E, Kimpel J, Herzog S, von Laer D. Heparan sulfate proteoglycans serve as alternative receptors for low affinity LCMV variants. PLoS Pathog. 2021 Oct 14;17(10):e1009996. doi: 10.1371/journal.ppat.1009996. eCollection 2021 Oct.
- Pipperger L, Riepler L, Kimpel J, Siller A, Stoitzner P, Bánki Z, von Laer D. Differential infection of murine and human dendritic cell subsets by oncolytic vesicular stomatitis virus variants. Oncoimmunology. 2021 Aug 31;10(1):1959140. doi:10.1080/2162402X.2021.1959140. eCollection
- Rössler A, Riepler L, Bante D, von Laer D, Kimpel J. SARS-CoV-2 Omicron Variant Neutralization in Serum from Vaccinated and Convalescent Persons. N Engl J Med. 2022 Feb 17;386(7):698-700. doi: 10.1056/NEJMc2119236. Epub 2022 Jan 12.
Selection of Funding
NIH – NIAID Save Variant Pipeline (15.08.2022 – 14.08.2023) – Dr. Janine Kimpel
FWF – SARS-CoV2 Kontrollierte-Kineasenfunktion (21.06.2021 – 21.06.2024) – Prof. Eduard Stefan / Dr. Janine Kimpel
Klinikum der Universität München – Präklinische Charakteriung von VSV-GP-HPV16 (01.11.2022 – 31.12.2025)– Dr. Janine Kimpel
EU – CleanAir (23.03.2021 – 31.01.2023) – Dr. Janine Kimpel / Prof. Dr. Dorothee von Laer
FWF – Inhibitionsprinzipien der SARS-CoV-2 Proteasen (15.09.2021 – 15.09.2024) – Emmanuel Heilmann, PhD / Prof. Dr. Dorothee von Laer
ViraT – Preclinical development (01.04.2021 – 31.03.2022, 01.04.2022 – 31.03.2023, 01.04.2023 – 31.03.2024) – Prof. Dr. Dorothee von Laer
Collaborations
Prof. Florian Krammer
Department of Microbiology
Icahn School of Medicine at Mount Sintai
USA
Prof. Reuben Stewart Harris
Department of Biochemistry & Structural Biology
University of Texas Health Science Center
Howard Hughes Medical Institute
USA
Prof. Luis Martinez-Sobrido
Texas Biomedical Research Institute
Department of MIMG, UTHSCSA
Department of Molecular Microbiology & Immunology, UTSA
USA
Prof. Stefano Moro
Molecular Modeling Section (MMS)
Department of Pharmaceutical and Pharmacological Sciences
University of Padova
Italy
MMag. Dr. Jörg Paetzold
Department of economics
University of Salzburg
Austria
Dr. Hannes Winner
Department of Economics
University of Salzburg
Austria
Prof. Derek J. Smith
Department of Zoology
University of Cambridge
UK
Prof. Dr. Ralf Wagner
Institute of Medical Microbiology and Hygiene
Molecular Microbiology (Virology)
University of Regensburg
Germany
PD DDr. Angelika Riemer
DKFZ
Ruprecht-Karls-Universität Heidelberg
Germany
PD Dr. Christof Geldmacher
Infection and tropical medicine
Ludwig Maximlian University of Munich
Germany
Daniela Weiskopf, PhD
La Jolla Institute for Immunology (LJI)
USA