Innrain 80
6020 Innsbruck
Email: Matthias.Erlacher@i-med.ac.at
Website: https://www.i-med.ac.at/rnomics/
Research year
Research Branch (ÖSTAT Classification)
106002, 106014, 106023,
Keywords
ncRNAs and diseases, Non-coding RNAs, ribosome translation, RNA modifications, and splicing mitochondrial tRNAs
Research Focus
All cells of all organisms contain two types of RNA molecules: messenger RNAs and so-called “non-protein-coding RNAs”. Many known ncRNAs are involved in the regulation of gene expression. Our research focuses on the identification of regulatory non-coding RNAs, their functional characterization and their roles in human diseases. In addition to working on regulatory RNAs, we are also studying the role of RNA modifications in mRNAs, tRNAs and rRNAs during protein synthesis.
General Facts
The Institute of Genomics and RNomics is home to two research groups that collaborate closely on various topics related to RNA function. The interests range from the identification and functional characterization of regulatory ncRNAs and their roles in human diseases to the roles of ncRNAs in splicing and in various aspects of protein synthesis and RNA modifications. The common goal is a better understanding of the numerous functions and roles of the classes of RNAs. The Hüttenhofer group is particularly interested in the characterization of ncRNAs that are involved in neurodegenerative diseases such as Alzheimer, in neurodevelopmental diseases such as Prader Willi syndrome or in the Schaaf-Yang syndrome. More recently, the group has also worked on mitochondrial genetic elements found in the nuclear genome. The Erlacher group is focusing on various aspects of protein synthesis, ranging from initiation to termination, and its regulation by modified RNAs. The projects are enabled by continuous third-party funding (e.g. FWF principal investigator projects, SFBs). The Institute is teaching molecular medicine (bachelor and master) and human medicine and is involved in the PhD curriculum. It regularly contributes to “Die lange Nacht der Forschung” and to “Open days” to introduce its research to the public and to encourage young pupils to study molecular medicine.
Research
Roles of ncRNAs in human diseases
Project leader: Alexander Hüttenhofer
Prader-Willi Syndrom and Schaaf-Yang Syndrome: elucidation of molecular mechanisms
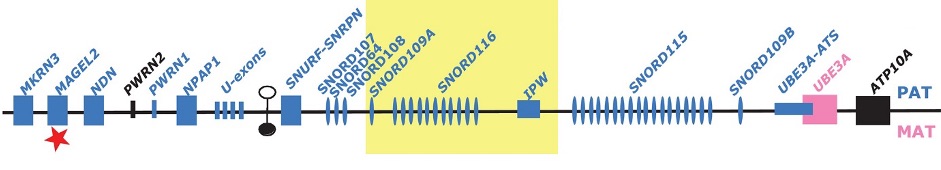
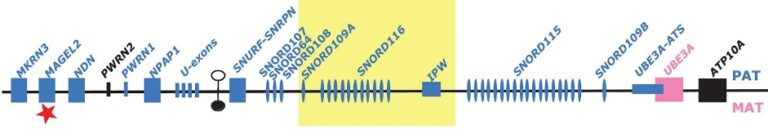
The genes of several brain-specific ncRNAs were first identified by the group by Experimental RNomics. They are in the class of small nucleolar RNAs (snoRNAs) located on chromosome 15q11-13 in humans (two of them, SNORD115 and SNRD116, in multi-copy repeats). The region is involved in the aetiology of the Prader-Willi-Syndrome (PWS), a neuro-developmental disease. We showed that the brain-specific RNAs are not present in PWS patients, pointing to a role of the RNAs in disease, as confirmed by the finding of microdeletions in PWS patients lacking the SNORD116 gene cluster. We are currently using molecular and cell biology approaches to investigate the role of brain-specific non-messenger RNAs in the aetiology of PWS.
ncRNAs and the SARS-Cov 2 virus infection
In collaboration with the Department of Hygiene and Microbiology (Prof. W. Posch), we are studying the small ncRNA transcriptome of lung epithelial cells infected with several variants of the SARS-Cov2 virus (e.g. delta and omicron variants). We prepared total RNA from non-infected and virus-infected lung epithelial cells and analysed the small ncRNA transcriptome of cellular ncRNAs and ncRNAs from the virus.
RNA modifications and their role in protein synthesis
Project leader: Matthias Erlacher
RNA modifications as regulators of translation
More than 100 modified nucleotides are known in various classes of RNA but, the biological role of many of them is not well understood. Taking advantage of developments in RNA chemistry, we are generating synthetic tRNAs and mRNAs that harbour different natural and non-natural modifications. The approach allows us to change single chemical groups within an RNA of interest. A systematic study of the modified molecules in a variety of experimental settings will provide a better understanding of essential processes such as initiation of translation, elongation and termination.
Superwobbling tRNAs
Superwobbling tRNAs are a specialized type of tRNA that extends the traditional wobble base pairing rules of protein synthesis
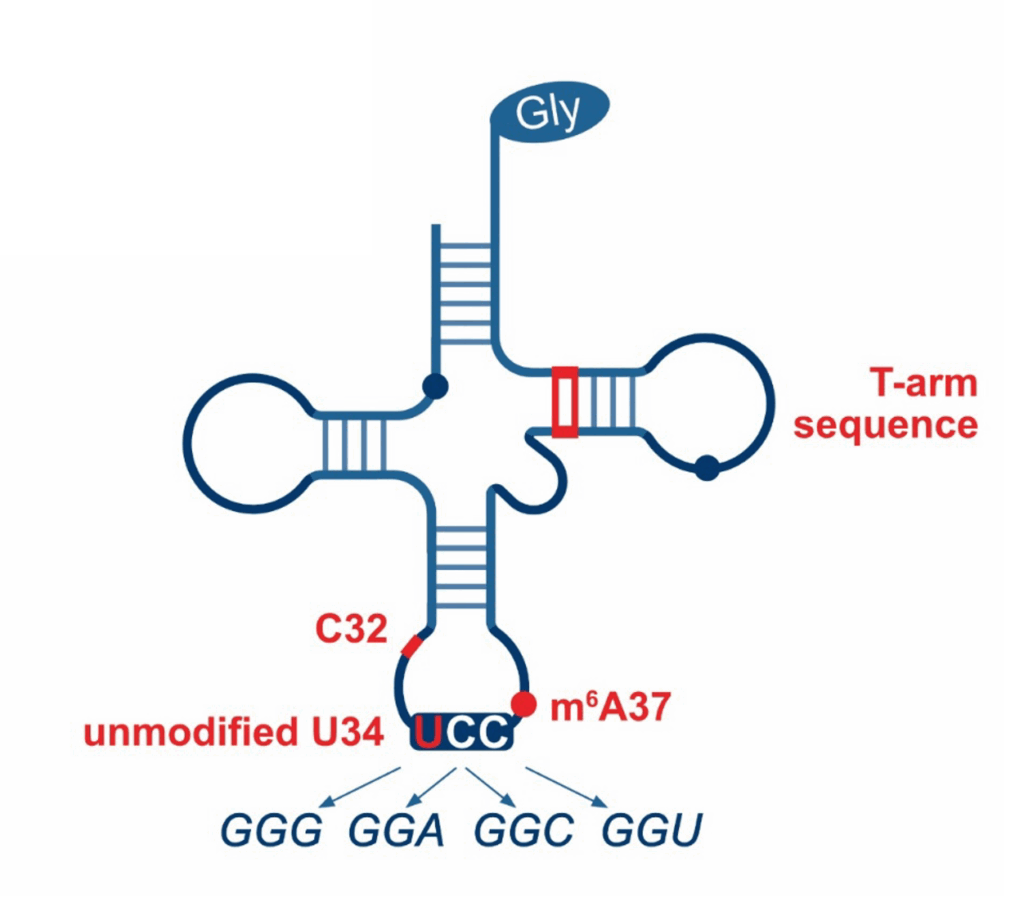
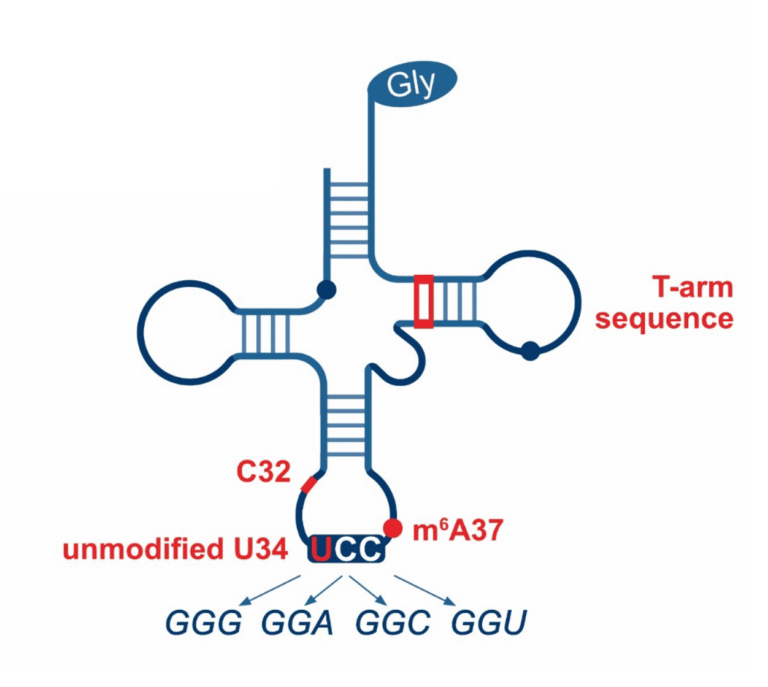
While conventional wobble base pairing allows a single tRNA to recognize multiple codons by less stringent base pairing at the third position, superwobbling enables a single tRNA to decode all synonymous codons. This decoding mechanism is largely restricted to specific genetic systems, including mitochondria, chloroplasts and several mycoplasma species. Although superwobbling has been known for decades, the exact molecular process that allows superwobbling tRNAs to expand their decoding capacity remains enigmatic. We are identifying the specific chemical groups and molecular residues that contribute to the unique decoding ability of these tRNAs.
Expanding the genetic code
A primary goal of synthetic biology is to expand the genetic code to overcome limitations in protein synthesis. The current genetic code, limited to 20 (21) canonical amino acids, constrains the potential diversity and functionality of proteins but the introduction of non-standard amino acids enables the engineering of proteins with unprecedented functions and properties. The key challenge is to create defined, unique codons that enable the site-specific incorporation of non-standard amino acids during translation. By modifying the tRNA anticodon and the mRNA codon, we aim to create novel, specific codon/anticodon interactions that enable the extension of the genetic code during translation.
Pictures
Selected Publications
Kohl, Maximilian P.; Kompatscher, Maria; Clementi, Nina; Holl, Lena; Erlacher, Matthias D.: Initiation at AUGUG and GUGUG sequences can lead to translation of overlapping reading frames in E. coli.
NUCLEIC ACIDS RESEARCH. 2023; 51(1); 271-289.
Kompatscher, Maria; Bartosik, Karolina; Erharter, Kevin; Plangger, Raphael; Juen, Fabian Sebastian; Kreutz, Christoph; Micura, Ronald; Westhof, Eric; Erlacher, Matthias D.: Contribution of tRNA sequence and modifications to the decoding preferences of E. coli and M. mycoides tRNAGlyUCC for synonymous glycine codons. NUCLEIC ACIDS RESEARCH. 2024; 52(3); 1374-1386.
Safari, MS.; Woerl, P.; Garmsiri, C.; Weber, D.; Kwiatkowski, M.; Hotze, M.; Kuenkel, L.; Lang, L.; Erlacher, M.; Gelpi, E.; Hainfellner, JA.; Baier, G.; Baier-Bitterlich, G.; Zur Nedden, S.: Glucose-1,6-bisphosphate: A new gatekeeper of cerebral mitochondrial pyruvate uptake.
MOLECULAR METABOLISM. 2024; 88; 102018.
Heimdoerfer, David; Vorleuter, Alexander; Eschlboeck, Alexander; Spathopoulou, Angeliki; Suarez-Cubero, Marta; Farhan, Hesso; Reiterer, Veronika; Spanjaard, Melanie; Schaaf, Christian P.; Huber, Lukas A.; Kremser, Leopold; Sarg, Bettina; Edenhofer, Frank; Geley, Stephan; de Araujo, Mariana E. G.; Huettenhofer, Alexander: Truncated variants of MAGEL2 are involved in the etiologies of the Schaaf-Yang and Prader-Willi syndromes. AMERICAN JOURNAL OF HUMAN GENETICS. 2024; 111(7);
Kompatscher, Maria; Gonnella, Isabell; Erlacher, Matthias D.: Studying the function of tRNA modifications: Experimental Challenges and opportunities. JOURNAL OF MOLECULAR BIOLOGY. 2025; 3:168934;
Selection of Funding
SFB F8004
Collaborations
Peter Stadler, Leipzig University, Germany
Heiner Schaal, Heinrich Heine University Düsseldorf, Germany
Utz Fischer, University of Würzburg, Germany
Eric Westhof, Université de Strasbourg, France
Ronald Micura, University of Innsbruck, Austria
Christoph Kreutz, University of Innsbruck, Austria