bis 2024: Univ.-Prof. Dr. med. Dorothee von Laer
Schöpfstraße 41
6020 Innsbruck
Fax: +43 (0)512 9003 73701
Email: Gisa.Gerold@i-med.ac.at
Website: https://virologie.i-med.ac.at/
Research year
Research Branch (ÖSTAT Classification)
301902, 301903, 301904, 301906, 302091, 303007, 303034, 106037, 106052
Keywords
antiviral therapies, cancer immunotherapy, clinical virology, host factors, immunogenic cell death, innate immunity, vaccines, virological diagnostic, Virology, virotherapy, and virus-host interaction
Research Focus
Improve prevention and treatment of viral infections
At the Institute of Virology, we strive to improve prevention and treatment of viral infections. Our diagnostic, research, training and knowledge transfer activities are geared towards developing modern virology and improving the quality of human life. In this endeavour, the section focuses on three key areas:
- Research on emerging virus – host interactions
- Development of antiviral therapies and vaccines
- Clinical virology including virological/serological diagnostics and viral oncotherapy.
General Facts
Structure:
The Institute of Virology has two professors: the Director Univ.-Prof. Dr. Gisa Gerold (Cellular Virology), and ao. Univ.-Prof. Dr. Heribert Stoiber (Molecular Virology). In addition, there are also three junior group leaders: Ass.Prof. PD Dr. Janine Kimpel (Infection Immunology and Vaccine Research), Dr. Wegene Borena (Clinical Virology/Epidemiology) and Dr. Zoltan Banki (Immunology of Virus Infection).
Clinical routine:
Approximately 30% of employees work in the serological and virological diagnostics group, servicing the University Hospital of Innsbruck (LKI), regional hospitals and medical practices in Tirol. During the first wave of the SARS-CoV-2 pandemic, the diagnostics laboratory was at the forefront of high-throughput PCR testing (over 1,000 tests per day) in Tyrol. Up to 200,000 tests per year are performed, covering all clinically relevant viruses. Apart from routine work, the diagnostics laboratory sets up tests for potential new viral threats as part of epidemic/pandemic preparedness.
Collaborations:
The Institute is involved in several international and national collaborative projects, a selection of which is mentioned below:
The Institute is leading the Human Frontier Science Programme project “Understanding fundamental mechanisms governing insect cell membrane deformability”, in collaboration with Harvard University, Boston, USA and University of Queensland, Brisbane, Australia.
The Institute is also part of a project funded by the National Health and Medical Research Council (NHMRC) with the Australian Berghofer Medical Research Institute.
The Institute is furthermore part of the European HIV Alliance (EHVA), which is funded by the European Union’s Horizon 2020 Research and Innovation Programme and collaborates with several international groups (p.8.) as well as with groups and clinics in Innsbruck: Haematology and Oncology (Wolf), Cell Genetics (Baier and Kleiter), Bioinformatics (Trajanoski), Pathophysiology (Geley), ENT (Riechelmann and Schartinger), Urology (Culig and Horninger) and Dermatology (Stoitzner) a.o.
The Institute has developed an oncolytic viral cancer vaccine, as well as complement-enhanced therapeutic antibodies. To bring these two developments to the clinic, two companies were founded: ViraTherapeutics GmbH (founded by D.v. Laer) and Lysovac (founder H. Stoiber), respectively. In September 2018, ViraTherapeutics was acquired by Boehringer Ingelheim.
Core facility:
The Institute established the BSL2 and BSL3 animal facilities of Innsbruck Medical University. These facilities include an in-vivo imaging system (IVIS PerkinElmer). The Institute has brought a state of the art high-resolution LC-MS machine to campus. It is used for internal and collaborative shotgun proteomics projects.
International boards
The Institute is active on international boards such as the Society for Virology (board of directors) and the WHO Technical Advisory Group on Composition of the Novel Corona virus vaccine (TAG-CO-VAC).
Research
Cellular Virology
Gisa Gerold
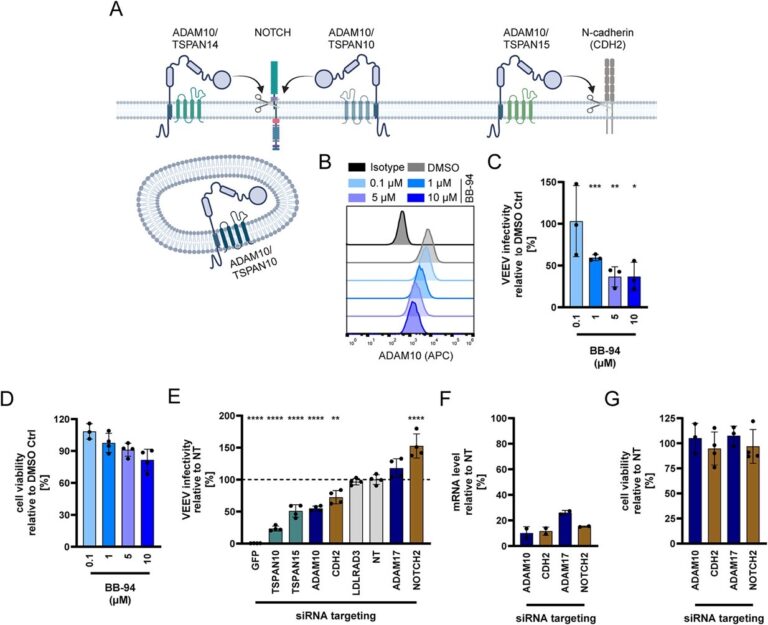
The emergence of viruses is a global challenge that is being accelerated by human activity. In this context, it is clear that climate change has led to the spread of virus-transmitting mosquito species across Europe. We investigate host protein networks involved in viral infection using re-emerging mosquito-borne viruses, such as the chikungunya virus, and emerging viruses, such as SARS-CoV-2. The group is using proteomics techniques for this purpose. We have identified the tetraspanin CD81 as host factor for the arthritis-like symptoms causing chikungunya virus and the metalloproteinase ADAM10 as a host factor for an encephalitis-causing alphavirus (Figure 1). This opens new avenues for the development of antiviral drugs and the understanding of host and tissue tropism. This will ultimately contribute to risk assessment of transmission routes, disease progression and the pandemic potential of viruses.
The group is replacing and reducing animal experimentation. It uses organoid and tissue explant models to dissect the intricate interplay of viruses and their host. We established skin explant models to mimic virus transmission from mosquitoes to humans, gut organoids to study gastrointestinal pathogens including norovirus and adenoviruses, and ultimately brain organoids to delineate how viruses cause encephalitis. These advanced models provide a comprehensive view of how viruses cause disease and are key to identifying important drug targets.
Infection Immunology and Vaccine Research
Janine Kimpel
The viral vector VSV-GP is a promising candidate as viral vector vaccine. It is a Chimeric Vesicular Stomatitis Virus (VSV) with the glycoprotein from the lymphocytic LCMV. We evaluated VSV-GP as an HIV vaccine by using next-generation Envelope antigens and combining the vector in heterologous prime/boost regimens with other viral or non-viral vaccine platforms. VSV-GP is also explored as a therapeutic vaccine against infections with the Human Papilloma Virus (HPV). The concept here is to induce a T cell response against HPV proteins and thereby eliminate infected (pre-) malignant cells.
New SARS-CoV-2 variants are emerging and will require updates to existing vaccines. It is vital to consider the basic antigenic relations of the variants and the neutralising antibody titres against these newly emerging variants in the general population when selecting a vaccine strain. Antigenic cartography is the tool we use to assess the need for vaccine updates. Repeated exposure generally increases cross-neutralisation, even against non-exposed variants. The number of exposures, antigenic relations and order of exposed variants influence neutralisation patterns.
Epidemiology
Wegene Borena
The large SARS-CoV-2 Beta variant outbreak in the district of Schwaz in spring 2021 was tackled with a rapid mass-immunisation campaign initiated and supported by the European Union. All adult inhabitants of the district were offered two doses of an mRNA vaccine. The Institute of Virology initiated an open-label, phase 4 trial to analyse immunity and breakthrough infections in the participants. SARS-CoV-2-specific immunity has been analysed one and seven months after the second dose, with participants performing weekly SARS-CoV-2 rapid antigen tests. This large observational study used a time-to-event approach to quantify the predictive value of vaccine-induced antibodies for the incidence of breakthrough infection. It confirmed these as correlates of protection.
Furthermore, we characterised antibody avidity, which defines the cumulative binding capacity of antibodies to the target antigen. The high binding affinity of neutralising or non-neutralising antibodies is clear evidence of the robustness of the induced immunity. The dynamics of avidity maturation following vaccination were investigated, comparing different vaccination regimens. Further investigation compared the robustness of infection-induced immune response to the vaccine-induced response in the context of antibody avidity.
Molecular Virology
Heribert Stoiber
Our research efforts have continued to elucidate the effect of the complement systems in various settings, such as viral pathogenesis, tumour immunology, and Periodontal Ehlers-Danlos syndrome (pEDS). pEDS is an autosomal dominant disorder. It is characterised by early-onset periodontitis. This leads to the premature loss of teeth, lack of attached gingiva and thin, fragile gums. These factors result in gingival recession. pEDS is caused by heterozygous missense mutations in the C1R and C1S genes of the classical complement C1 complex. Our results show that the pathogenesis of pEDS is not solely mediated by activation of the complement cascade but by inadequate C1s-mediated degradation of matrix proteins, confirming pEDS as a primary connective tissue disorder. Antibody-mediated complement-dependent cytotoxicity (CDC) is vital for the elimination of chronic lymphocytic leukaemia (CLL) cells. CDC was further enhanced by directly fusing the Factor H subunits SCR1920 to monoclonal antibodies targeting CLL cells. We also showed that severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) hijacks CD55 and CD59, two members of the host cellular Regulators of Complement Activation (RCA) family, as well as serum-derived Factor H to resist antibody-dependent complement-mediated lysis triggered by sera from immunised individuals. Blocking the biological functions of virion-associated CD55 and CD59 and competing for Factor H recruitment with functionally inactive recombinant Factor H-derived short consensus repeats (SCR18-20) restored SARS-CoV-2 complement sensitivity in a synergistic manner.
Immunology of Virus Infection
Zoltan Banki
Oncolytic virotherapy is a promising therapeutic strategy for cancer. Such viruses replicate selectively in tumour cells, destroying them while activating anti-tumour immune responses. VSV-GP, a chimeric vesicular stomatitis virus variant with the glycoprotein of the lymphocytic choriomeningitis virus, is a potent oncolytic virus. Our preclinical studies will optimise their efficacy for future clinical applications by enhancing and sustaining tumour-specific T cell responses.
We are also interested in antiviral T cell immunity in the context of viral infections and vaccinations. We focus particularly on respiratory pathogens such as SARS-CoV-2 and respiratory syncytial virus (RSV). We study the dynamics of T cell activation, memory formation, and cytokine production. This allows us to better understand the mechanisms that govern protective immunity and immunopathology. We are integrating cutting-edge techniques like spectral flow cytometry to unravel the complexities of virus-host interactions and contribute to the development of next-generation antiviral therapies, vaccines, and immunotherapies.
Viral-based tools to predict antiviral resistance
Dorothee von Laer (Francesco Costacurta, Emmanuel Heilmann)
We built on our previous work using a VSV-based tool to predict and test mutations of SARS-CoV-2 main protease (Mpro) against the protease inhibitor nirmatrelvir (Paxlovid) (Figure 2A). A mutation selection workflow coupled with next-generation-sequencing (Nanopore) has been implemented, facilitating the processing of larger numbers of samples (Figure 2B) (PMID: 39259728). Improving the initial method allowed us to perform similar studies with ensitrelvir, another clinically approved Mpro inhibitor (PMID: 39053514), and with the MERS-CoV main protease (PMID: 38933182). We have established collaborations with several groups. These will support and cross-validate our findings. They will also provide potential molecular mechanisms of resistance by performing orthogonal in vitro assays and in silico/molecular modelling studies, respectively.
We adapted this system and developed a dual-chimeric VSV-Spike-Mpro (Figure 2C) to generate, predict and test mutations of the SARS-CoV-2 Spike with reduced susceptibility to an entry/fusion inhibitor developed by RedQueen Therapeutics. Such surrogate systems represent a safe gain of function technology and are critical to assess the potential of drug resistance development.
Pictures
Selected Publications
Carriquí-Madroñal B, Lasswitz L, von Hahn T, Gerold G. Genetic and pharmacological perturbation of hepatitis-C virus entry. Curr Opin Virol. 2023 Oct;62:101362. doi: 10.1016/j.coviro.2023.101362. Epub 2023 Sep 6. PMID: 37678113.
Carriquí-Madroñal B, Sheldon J, Duven M, Stegmann C, Cirksena K, Wyler E, Zapatero-Belinchón FJ, Vondran FWR, Gerold G. The matrix metalloproteinase ADAM10 supports hepatitis C virus entry and cell-to-cell spread via its sheddase activity. PLoS Pathog. 2023 Nov 15;19(11):e1011759. doi: 10.1371/journal.ppat.1011759. PMID: 37967063; PMCID: PMC10650992.
Alvarez KG, Goral L, Suwandi A, Lasswitz L, Zapatero-Belinchón FJ, Ehrhardt K, Nagarathinam K, Künnemann K, Krey T, Wiedemann A, Gerold G, Grassl GA. Human tetraspanin CD81 facilitates invasion of Salmonella enterica into human epithelial cells. Virulence. 2024 Dec;15(1):2399792. doi:10.1080/21505594.2024.2399792. Epub 2024 Sep 24. PMID: 39239914; PMCID: PMC11423668.
Dos Reis VP, Cirksena K, Rybak-Wolf A, Seeger B, Herker E, Gerold G. 3D Spheroid and Organoid Models to Study Neuroinfection of RNA Viruses. Methods Mol Biol. 2024;2824:409-424. doi: 10.1007/978-1-0716-3926-9_26. PMID: 39039427.
Haid S, Matthaei A, Winkler M, Sake SM, Gunesch AP, Milke V, Köhler NM, Rückert J, Vieyres G, Kühl D, Nguyen T-T, Göhl M, Lasswitz L, Zapatero-Belinchón FJ, Brogden G, Gerold G, Wiegmann B, Bilitewski U, Brown RJP, Brönstrup M, Schulz TF, Pietschmann T. Repurposing screen identifies novel candidates for broad-spectrum coronavirus antivirals and druggable host targets. Antimicrob Agents Chemother. 2024 Mar 6;68(3):e0121023. doi: 10.1128/aac.01210-23. Epub 2024 Feb 6. PMID: 38319076; PMCID: PMC10916382.
Matthaei A, Joecks S, Frauenstein A, Bruening J, Bankwitz D, Friesland M, Gerold G, Vieyres G, Kaderali L, Meissner F, Pietschmann T. Landscape of protein-protein interactions during hepatitis C virus assembly and release. Microbiol Spectr. 2024 Feb 6;12(2):e0256222. doi: 10.1128/spectrum.02562-22. Epub 2024 Jan 17. PMID: 38230952; PMCID: PMC10846047.
Bley H, Krisp C, Schöbel A, Hehner J, Schneider L, Becker M, Stegmann C, Heidenfels E, Nguyen-Dinh V, Schlüter H, Gerold G, Herker E. Proximity labeling of host factor ANXA3 in HCV infection reveals a novel LARP1 function in viral entry. J Biol Chem. 2024 May;300(5):107286. doi: 10.1016/j.jbc.2024.107286. Epub 2024 Apr 16. PMID: 38636657; PMCID: PMC11101947.
Becker M, Conca DV, Dorma N, Mistry N, Hahlin E, Frängsmyr L, Bally M, Arnberg N, Gerold G. Efficient clathrin-mediated entry of enteric adenoviruses in human duodenal cells. J Virol. 2023 Oct 31;97(10):e0077023. doi: 10.1128/jvi.00770-23. Epub 2023 Oct 12. PMID: 37823645; PMCID: PMC10617564.
Wilken L, Lasswitz L, Scaturro P, Gerold G. Identification of RVFV Host Factors Using Quantitative Interaction Proteomics. Methods Mol Biol. 2024;2824:189-202. doi: 10.1007/978-1-0716-3926-9_13. PMID: 39039414.
Löw K, Möller R, Stegmann C, Becker M, Rehburg L, Obernolte H, Schaudien D, Oestereich L, Braun A, Kunz S, Gerold G. Luminescent reporter cells enable the identification of broad-spectrum antivirals against emerging viruses. J Med Virol. 2023 Nov;95(11):e29211. doi: 10.1002/jmv.29211. PMID: 37975336.
Victoria C, Schulz G, Klöhn M, Weber S, Holicki CM, Brüggemann Y, Becker M, Gerold G, Eiden M, Groschup MH, Steinmann E, Kirschning A. Halogenated Rocaglate Derivatives: Pan-antiviral Agents against Hepatitis E Virus and Emerging Viruses. J Med Chem. 2024 Jan 11;67(1):289-321. doi: 10.1021/acs.jmedchem.3c01357. Epub 2023 Dec 21. PMID: 38127656; PMCID: PMC10788925.
Li D, Bühler M, Runft S, Gerold G, Marek K, Baumgärtner W, Strowig T, Gerhauser I. ASC- and caspase-1-deficient C57BL/6 mice do not develop demyelinating disease after infection with Theiler’s murine encephalomyelitis virus. Sci Rep. 2023 Jul 6;13(1):10960. doi: 10.1038/s41598-023-38152-3. PMID:37414913; PMCID: PMC10326010.
Passos V, Henkel LM, Wang J, Zapatero-Belinchón FJ, Möller R, Sun G, Waltl I, Schneider T, Wachs A, Ritter B, Kropp KA, Zhu S, Deleidi M, Kalinke U, Schulz TF, Höglinger G, Gerold G, Wegner F, Viejo-Borbolla A. Innate immune response to SARS-CoV-2 infection contributes to neuronal damage in human iPSC-derived peripheral neurons. J Med Virol. 2024 Feb;96(2):e29455. doi: 10.1002/jmv.29455. PMID: 38323709.
Mhlekude B, Postmus D, Stenzel S, Weiner J 3rd, Jansen J, Zapatero-Belinchón FJ, Olmer R, Richter A, Heinze J, Heinemann N, Mühlemann B, Schroeder S, Jones TC, Müller MA, Drosten C, Pich A, Thiel V, Martin U, Niemeyer D, Gerold G, Beule D, Goffinet C. Pharmacological inhibition of bromodomain and extra-terminal proteins induces an NRF-2-mediated antiviral state that is subverted by SARS-CoV-2 infection. PLoS Pathog. 2023 Sep 25;19(9):e1011657. doi: 10.1371/journal.ppat.1011657. PMID: 37747932; PMCID: PMC10629670.
Marek K, Armando F, Asawapattanakul T, Nippold VM, Plattet P, Gerold G, Baumgärtner W, Puff C. Functional Granulocyte-Macrophage Colony-Stimulating Factor (GM-CSF) Delivered by Canine Histiocytic Sarcoma Cells Persistently Infected with Engineered Attenuated Canine Distemper Virus. Pathogens. 2023 Jun 27;12(7):877. doi: 10.3390/pathogens12070877. PMID: 37513724; PMCID:PMC10385001.
Costacurta F, Dodaro A, Bante D, Schöppe H, Peng JY, Sprenger B, He X, Moghadasi SA, Egger LM, Fleischmann J, Pavan M, Bassani D, Menin S, Rauch S, Krismer L, Sauerwein A, Heberle A, Rabensteiner T, Ho J, Harris RS, Stefan E, Schneider R, Dunzendorfer-Matt T, Naschberger A, Wang D, Kaserer T, Moro S, von Laer D, Heilmann E. A comprehensive study of SARS-CoV-2 main protease (Mpro) inhibitor-resistant mutants selected in a VSV-based system. PLoS Pathog. 2024 Sep 11;20(9):e1012522. doi: 10.1371/journal.ppat.1012522. PMID: 39259728; PMCID: PMC11407635.
Rauch S, Costacurta F, Schöppe H, Peng JY, Bante D, Erisoez EE, Sprenger B, He X, Moghadasi SA, Krismer L, Sauerwein A, Heberle A, Rabensteiner T, Wang D, Naschberger A, Dunzendorfer-Matt T, Kaserer T, von Laer D, Heilmann E. Highly specific SARS-CoV-2 main protease (Mpro) mutations against the clinical antiviral ensitrelvir selected in a safe, VSV-based system. Antiviral Res. 2024 Nov;231:105969. doi: 10.1016/j.antiviral.2024.105969. Epub 2024 Jul 23. PMID: 39053514.
Borena W, Kitchen M, Gisinger M, Taylor N, Oberkofler H, Dewasurendra D, Widschwendter A, Stoiber H, von Laer D, Sarcletti M. Disproportionate preponderance of HPV genotypes associated with anogenital warts among HIV-positive MSM. Front Public Health. 2024 Sep 20;12:1437309. doi:10.3389/fpubh.2024.1437309. PMID: 39371203; PMCID: PMC11449850.
Rössler A, Knabl L, Raschbichler LM, Peer E, von Laer D, Borena W, Kimpel J. Reduced sensitivity of antibody tests after omicron infection. Lancet Microbe. 2023 Jan;4(1):e10-e11. doi: 10.1016/S2666-5247(22)00222-1. Epub 2022 Sep 19. PMID: 36137554; PMCID: PMC9484764.
Moghadasi SA, Heilmann E, Khalil AM, Nnabuife C, Kearns FL, Ye C, Moraes SN, Costacurta F, Esler MA, Aihara H, von Laer D, Martinez-Sobrido L, Palzkill T, Amaro RE, Harris RS. Transmissible SARS-CoV-2 variants with resistance to clinical protease inhibitors. Sci Adv. 2023 Mar 29;9(13):eade8778. doi: 10.1126/sciadv.ade8778. Epub 2023 Mar 29. PMID: 36989354; PMCID: PMC10058310.
Rössler A, Netzl A, Knabl L, Bante D, Wilks SH, Borena W, von Laer D, Smith DJ, Kimpel J. Characterizing SARS-CoV-2 neutralization profiles after bivalent boosting using antigenic cartography. Nat Commun. 2023 Aug 26;14(1):5224. doi: 10.1038/s41467-023-41049-4. PMID: 37633965; PMCID: PMC10460376.
Runge A, Straif S, Banki Z, Borena W, Muellauer B, Brunner J, Gottfried T, Schmutzhard J, Dudas J, Risslegger B, Randhawa A, Lass-Flörl C, von Laer D, Riechelmann H. Viral infection in chronic otitis media with effusion in children. Front Pediatr. 2023 May 10;11:1124567. doi: 10.3389/fped.2023.1124567. PMID: 37234860; PMCID: PMC10208354.
Heilmann E, Costacurta F, Moghadasi SA, Ye C, Pavan M, Bassani D, Volland A, Ascher C, Weiss AKH, Bante D, Harris RS, Moro S, Rupp B, Martinez-Sobrido L, von Laer D. SARS-CoV-2 3CLpro mutations selected in a VSV-based system confer resistance to nirmatrelvir, ensitrelvir, and GC376. Sci Transl Med. 2023 Jan 11;15(678):eabq7360. doi: 10.1126/scitranslmed.abq7360. Epub 2023 Jan 11. PMID: 36194133; PMCID: PMC9765458.
Riepler L, Frommelt LS, Wilmschen-Tober S, Mbuya W, Held K, Volland A, von Laer D, Geldmacher C, Kimpel J. Therapeutic Efficacy of a VSV-GP-based Human Papilloma Virus Vaccine in a Murine Cancer Model. J Mol Biol. 2023 Jul 1;435(13):168096. doi: 10.1016/j.jmb.2023.168096. Epub 2023 Apr 20. PMID:37086948.
Schuler LM, Falkensammer B, Orlik P, Auckenthaler M, Kranewitter C, Bante D, von Laer D, Fink FM. Terminal Ileitis as the Exclusive Manifestation of COVID-19 in Children. Microorganisms. 2024 Jul 6;12(7):1377. doi: 10.3390/microorganisms12071377. PMID: 39065145; PMCID: PMC11279043.
Rauch S, Costacurta F, von Laer D, Heilmann E. Vesicular Stomatitis Virus as a Platform for Protease Activity Measurements. Curr Protoc. 2024 Nov;4(11):e70062. doi: 10.1002/cpz1.70062. PMID: 39570195; PMCID: PMC11580764.
Schatz C, Knabl L, Lee HK, Seeboeck R, von Laer D, Lafon E, Borena W, Mangge H, Prüller F, Qerimi A, Wilflingseder D, Posch W, Haybaeck J. Machine Learning to Identify Critical Biomarker Profiles in New SARS-CoV-2 Variants. Microorganisms. 2024 Apr 15;12(4):798. doi: 10.3390/microorganisms12040798. PMID: 38674742; PMCID: PMC11052335.
Costacurta F, Dodaro A, Bante D, Schöppe H, Sprenger B, Moghadasi SA, Fleischmann J, Pavan M, Bassani D, Menin S, Rauch S, Krismer L, Sauerwein A, Heberle A, Rabensteiner T, Ho J, Harris RS, Stefan E, Schneider R, Kaserer T, Moro S, von Laer D, Heilmann E. A comprehensive study of SARS-CoV-2 main
Protease (Mpro) inhibitor-resistant mutants selected in a VSV-based system. bioRxiv [Preprint]. 2023 Oct 4:2023.09.22.558628. doi: 10.1101/2023.09.22.558628. Update in: PLoS Pathog. 2024 Sep 11;20(9):e1012522. doi: 10.1371/journal.ppat.1012522. PMID: 37808638; PMCID: PMC10557589.
Rössler A, Knabl L, Netzl A, Bante D, Borena W, von Laer D, Smith DJ, Kimpel J. Durability of Cross-Neutralizing Antibodies 5.5 Months After Bivalent Coronavirus Disease 2019 Vaccine Booster. J Infect Dis. 2024 Mar 14;229(3):644-647. doi: 10.1093/infdis/jiad472. PMID: 38016020; PMCID: PMC10938204.
Rössler A, Netzl A, Knabl L, Wilks SH, Mühlemann B, Türeli S, Mykytyn A, von Laer D, Haagmans BL, Smith DJ, Kimpel J. Direct comparison of SARS-CoV-2 variant specific neutralizing antibodies in human and hamster sera. NPJ Vaccines. 2024 May 18;9(1):85. doi: 10.1038/s41541-024-00888-y. PMID: 38762525; PMCID: PMC11102554.
Harthaller T, Falkensammer B, Bante D, Huber M, Schmitt M, Benainouna H, Rössler A, Fleischer V, von Laer D, Kimpel J, Wuerzner R, Borena W. Retained avidity despite reduced cross-binding and cross-neutralizing antibody levels to Omicron after SARS-COV-2 wild-type infection or mRNA double vaccination. Front Immunol. 2023 Jul 21;14:1196988. doi: 10.3389/fimmu.2023.1196988. PMID:37545492; PMCID: PMC10401431.
Perdiguero B, Hauser A, Gómez CE, Peterhoff D, Sideris E, Sorzano CóS, Wilmschen S, Schaber M, Stengel L, Asbach B, Ding S, Von Laer D, Levy Y, Pantaleo G, Kimpel J, Esteban M, Wagner R. Potency and durability of T and B cell immune responses after homologous and heterologous vector delivery of a trimer-stabilized, membrane-displayed HIV-1 clade ConC Env protein. Front Immunol. 2023 Nov 17;14:1270908. doi: 10.3389/fimmu.2023.1270908. PMID: 38045703; PMCID: PMC10690772.
Seekircher L, Banki Z, Kimpel J, Rössler A, Schäfer H, Falkensammer B, Bante D, Forer L, Schönherr S; Shieldvacc-2 Study Group; Harthaller T, Sacher M, Ower C, Tschiderer L, Ulmer H, Krammer F, von Laer D, Borena W, Willeit P. Immune response after two doses of the BNT162b2 COVID-19 vaccine and risk of SARS-CoV-2 breakthrough infection in Tyrol, Austria: an open-label, observational phase 4 trial. Lancet Microbe. 2023 Aug;4(8):e612-e621. doi: 10.1016/S2666-5247(23)00107-6. Epub 2023 Jun 21. PMID: 37354911; PMCID: PMC10284585.
Costacurta F, Rauch S, von Laer D, Heilmann E. Using Vesicular Stomatitis Virus as a Platform for Directed Protease Evolution. Curr Protoc. 2024 Dec;4(12):e70074. doi: 10.1002/cpz1.70074. PMID: 39711492; PMCID: PMC11664493.
Kimpel J, Rössler A, Bante D, Borena W, von Laer D, Zehetner C, Rauchegger T, Seiwald S, Falkensammer B. Detection of infectious SARS-CoV-2 in ocular samples is linked to viral load in the nasopharynx. Front Cell Infect Microbiol. 2024 Mar 4;14:1332157. doi: 10.3389/fcimb.2024.1332157. PMID: 38500504; PMCID: PMC10946250.
Krismer L, Schöppe H, Rauch S, Bante D, Sprenger B, Naschberger A, Costacurta F, Fürst A, Sauerwein A, Rupp B, Kaserer T, von Laer D, Heilmann E. Study of key residues in MERS-CoV and SARS-CoV-2 main proteases for resistance against clinically applied inhibitors nirmatrelvir and ensitrelvir. Npj Viruses.
2024;2(1):23. doi: 10.1038/s44298-024-00028-2. Epub 2024 Jun 24. Erratum in: Npj Viruses. 2025 Mar 5;3(1):18. doi: 10.1038/s44298-025-00094-0. PMID: 38933182; PMCID: PMC11196219.
Gebetsberger L, Malekshahi Z, Teutsch A, Tajti G, Fontaine F, Marella N, Mueller A, Prantl L, Stockinger H, Stoiber H, Ohradanova-Repic A. SARS-CoV-2 hijacks host CD55, CD59 and factor H to impair antibody-dependent complement-mediated lysis. Emerg Microbes Infect. 2024 Dec;13(1):2417868. doi: 10.1080/22221751.2024.2417868. Epub 2024 Oct 28. PMID: 39435487; PMCID: PMC11520101.
Prantl L, Heider P, Bergmeister L, Calana K, Bohn JP, Wolf D, Banki Z, Bosch A, Plach M, Huber G, Schrödel S, Thirion C, Stoiber H. Enhancement of complement-dependent cytotoxicity by linking factor-H derived short consensus repeats 19-20 to CD20 antibodies. Front Immunol. 2024 Jul 22;15:1379023. doi: 10.3389/fimmu.2024.1379023. PMID: 39104533; PMCID: PMC11298693.
Amberger A, Pertoll J, Traunfellner P, Kapferer-Seebacher I, Stoiber H, Klimaschewski L, Thielens N, Gaboriaud C, Zschocke J. Degradation of collagen I by activated C1s in periodontal Ehlers-Danlos Syndrome. Front Immunol. 2023 Mar 7;14:1157421. doi: 10.3389/fimmu.2023.1157421. PMID: 36960056; PMCID: PMC10028100.
Rosales A, Kuppelwieser S, Giner T, Hofer J, Riedl Khursigara M, Orth-Höller D, Borena W, Cortina G, Jungraithmayr T, Wuerzner R; German-Austrian H. U. S.-Study Group. Outcome 10 years after Shiga toxin-producing E. coli (STEC)-associated hemolytic uremic syndrome: importance of long-term follow-up. Pediatr Nephrol. 2024 Aug;39(8):2459-2465. doi: 10.1007/s00467-024-06355-z. Epub 2024 Apr 9. PMID: 38589699; PMCID: PMC11199238.
Kofler B, Widschwendter A, Hofauer B, Gatt C, Fabel S, Leichtle A, Ciresa-König A, Dudas J, Borena W. Is an oropharyngeal HPV infection more frequently detectable in women with a genital HPV infection? Eur Arch Otorhinolaryngol. 2024 Feb;281(2):1041-1046. doi: 10.1007/s00405-023-08314-0. Epub 2023 Nov 10. PMID: 37947818.
Kitchen M, Borena WT, Gisinger M, Meindl E, Wanner M, Govrins MA, Sarcletti M. Pharyngeal gonococcal infection and the sensitivity of oral gargle samples in comparison to self-collected throat swabs for the detection of N. gonorrhoeae in persons in Tyrol, Austria. Infection. 2025 Apr;53(2):547-552. doi: 10.1007/s15010-024-02359-x. Epub 2024 Aug 2. PMID: 39093382.
Becker B, Wottawa F, Bakr M, Koncina E, Mayr L, Kugler J, Yang G, Windross SJ, Neises L, Mishra N, Harris D, Tran F, Welz L, Schwärzler J, Banki Z, Stengel ST, Ito G, Krötz C, Coleman OI, Jaeger C, Haller D, Paludan SR, Blumberg R, Kaser A, Cicin-Sain L, Schreiber S, Adolph TE, Letellier E, Rosenstiel P, Meiser
J, Aden K. Serine metabolism is crucial for cGAS-STING signaling and viral defense control in the gut. iScience. 2024 Feb 8;27(3):109173. doi: 10.1016/j.isci.2024.109173. PMID: 38496294; PMCID: PMC10943449.
Fischhuber K, Banki Z, Kimpel J, Kragl N, Rössler A, Bolze A, Muellauer B, Angerer J, Nagy G, Nagy E, Szijarto V. Antiviral Potential of Azelastine against Major Respiratory Viruses. Viruses. 2023 Nov 23;15(12):2300. doi: 10.3390/v15122300. PMID: 38140540; PMCID: PMC10747764.
Loacker L, Kimpel J, Banki Z, Schmidt CQ, Griesmacher A, Anliker M. Increased PD-L1 surface expression on peripheral blood granulocytes and monocytes after vaccination with SARS-CoV2 mRNA or vector vaccine. Clin Chem Lab Med. 2022 Oct 18;61(1):e17-e19. doi: 10.1515/cclm-2022-0787. PMID: 36245120.
Muehlemann B, Wilks SH, Baracco L, Bekliz M, Carreño JM, Corman VM, Davis-Gardner ME, Dejnirattisai W, Diamond MS, Douek DC, Drosten C, Eckerle I, Edara VV, Ellis M, Fouchier RAM, Frieman M, Godbole S, Haagmans B, Halfmann PJ, Henry AR, Jones TC, Katzelnick LC, Kawaoka Y, Kimpel J, Krammer F, Lai L, Liu C, Lusvarghi S, Meyer B, Mongkolsapaya J, Montefiori DC, Mykytyn A, Netzl A, Pollett S, Rössler A, Screaton GR, Shen X, Sigal A, Simon V, Subramanian R, Supasa P, Suthar MS, Türeli S, Wang W, Weiss CD, Smith DJ. Comparative analysis of SARS-CoV-2 neutralization titers reveals consistency between human and animal model serum and across assays. Sci Transl Med. 2024 May 15;16(747):eadl1722.
doi: 10.1126/scitranslmed.adl1722. Epub 2024 May 15. PMID: 38748773.
Brangel P, Tureli S, Mühlemann B, Liechti N, Zysset D, Engler O, Hunger-Glaser I, Ghiga I, Mattiuzzo G, Eckerle I, Bekliz M, Rössler A, Schmitt MM, Knabl L, Kimpel J, Tort LFL, de Araujo MF, de Oliveira ACA, Caetano BC, Siqueira MM, Budt M, Gensch JM, Wolff T, Hassan T, Selvaraj FA, Hermanus T, Kgagudi P,
Crowther C, Richardson SI, Bhiman JN, Moore PL, Cheng SMS, Li JKC, Poon LLM, Peiris M, Corman VM, Drosten C, Lai L, Hunsawong T, Rungrojcharoenkit K, Lohachanakul J, Sigal A, Khan K, Thiel V, Barut GT, Ebert N, Mykytyn AZ, Owusu Donkor I, Aboagye JO, Nartey PA, Van Kerkhove MD, Cunningham J, Haagmans BL, Suthar MS, Smith D, Subissi L. A Global Collaborative Comparison of SARS-CoV-2 Antigenicity Across 15 Laboratories. Viruses. 2024 Dec 18;16(12):1936. doi: 10.3390/v16121936. PMID: 39772242; PMCID: PMC11680265.
Huang A, Riepler L, Rieder D, Kimpel J, Lusser A. No evidence for epitranscriptomic m5C modification of SARS-CoV-2, HIV and MLV viral RNA. RNA. 2023 Jun;29(6):756-763. doi: 10.1261/rna.079549.122. Epub 2023 Mar 8. Erratum in: RNA. 2024 Nov 18;30(12):1686. doi: 10.1261/rna.080282.124. PMID: 36889928; PMCID: PMC10187675.
Muehlemann B, Wilks SH, Baracco L, Bekliz M, Carreño JM, Corman VM, Davis-Gardner ME, Dejnirattisai W, Diamond MS, Douek DC, Drosten C, Eckerle I, Edara VV, Ellis M, Fouchier RAM, Frieman M, Godbole S, Haagmans B, Halfmann PJ, Henry AR, Jones TC, Katzelnick LC, Kawaoka Y, Kimpel J, Krammer F, Lai L, Liu C, Lusvarghi S, Meyer B, Mongkolsapaya J, Montefiori DC, Mykytyn A, Netzl A, Pollett S, Rössler A, Screaton GR, Shen X, Sigal A, Simon V, Subramanian R, Supasa P, Suthar M, Türeli S, Wang W, Weiss CD, Smith DJ. Comparative Analysis of SARS-CoV-2 Antigenicity across Assays and in Human and Animal Model Sera. bioRxiv [Preprint]. 2023 Sep 27:2023.09.27.559689. doi: 10.1101/2023.09.27.559689. PMID: 37808679; PMCID: PMC10557678.
Watschinger C, Stampfel G, Zollner A, Hoog AM, Rössler A, Reiter S, Dax K, Kimpel J, Tilg H, Antlanger M, Schwaiger E, Moschen AR. B and T Cell Responses to SARS-CoV-2 Vaccination in Kidney and Liver Transplant Recipients with and without Previous COVID-19. Viruses. 2023 Dec 19;16(1):1. doi: 10.3390/v16010001. PMID: 38275936; PMCID: PMC10820906.
Zollner A, Koch R, Jukic A, Pfister A, Meyer M, Wick N, Wick G, Rössler A, Kimpel J, Adolph TE, Tilg H. Clearance of Gut Mucosal SARS-CoV-2 Antigens and Postacute COVID-19 After 2 Years in Patients With Inflammatory Bowel Disease. Gastroenterology. 2024 Aug;167(3):604-607.e8. doi: 10.1053/j.gastro.2024.04.008. Epub 2024 Apr 16. PMID: 38631418.
Tschiderer L, Innerhofer H, Seekircher L, Waltle L, Richter L, Kimpel J, Lass-Flörl C, Forer L, Schönherr S, Larsen DA, Krammer F, Embacher-Aichhorn S, Tilg H, Weiss G, Allerberger F, Willeit P. Long-term effectiveness of an ultra-rapid rollout vaccination campaign with BNT162b2 on the incidence of SARS-CoV-2 infection. iScience. 2024 Oct 10;27(11):111117. doi: 10.1016/j.isci.2024.111117. PMID: 39555399; PMCID: PMC11567098.
Selection of Funding
Gisa Gerold
1. NHMRC Ideas Grant (01.01.2025 – 31.12.2027)
2. Swedish Research Council Grant (01.01.2025 – 31.12.2028)
3. HFSP funding (01.10.2023-31.12.2026)
4. DFG GE 2145/6-1 (01.11.2024 – 31.12.2026)
5. DFG INST 193/90-1 FUGG (DFG 50%; Landesmittel 45%; TiHo 5%) Forschungsgroßgeräteförderung
6. DFG VIPER (01.04.2025 – 31.03.2028
7. DFG Walter Benjamin funding (01.11.2024 – 31.10.2026)
8. BMBF MOZART (15.10.2023 – 30.06.2025)
9. JCK22-0047 Kempe Foundation (01.07.2023 – 31.12.2025)
10. Spitzenforschung für Niedersachen (MWK), Europa-Programm 15-76251-1-2/23 (511/2023) Förderlinie KONSORT (01.02.2023 – 01.05.2024)
Janine Kimpel
NIH – SAVE Concept Funding (15.09.2023 – 15.09.2024)
Collaborations
Prof. Dr. Norbert Perrimon
Norbert Perrimon Lab, Harvard Medical School, USA
Dr. Nick Ariottti,
Centre for Cell Biology of Chronic Disease, University of Queensland, Brisbane, Australia
Prof. Dr. Anna Oeverby-Wernstedt,
Department of Clinical Microbiology, Umea University, Sweden
Prof. Dr. Max Lenz,
Institute for Neuroanatomy and Cell Biology, Hannover Medical School, Germany
Prof. Dr. Florian Krammer
Department of Microbiology, Icahn School of Medicine at Mount Sinai, New York, USA & Semmelweis Institute, Vienna
Prof. Derek J. Smith
Department of Zoology, University of Cambridge, UK
Prof. Dr. Ralf Wagner
Institute of Medical Microbiology and Hygiene, Molecular Microbiology (Virology), University of Regensburg, Germany