Innrain 80-82
6020 Innsbruck
Fax: +43 512900373100
Email: Hubertus.haas@i-med.ac.at
Website: https://www.i-med.ac.at/molbio/
Research year
Research Branch (ÖSTAT Classification)
106023, 106022, 106024, 301114, 303020
Keywords
bioactive fungal products, Chromatin and epigenetics, drug resistance, filamentous fungi, fungal infection, iron metabolism, RNA epitranscriptomics, and siderophores and antimicrobial proteins
Research Focus
Fungal Metabolism, Gene Regulation, Virulence and Drug Resistance
- Iron metabolism in filamentous fungi: links to human disease
- Histone-modifying enzymes in gene regulation, in fungal physiology and as antifungal targets
- Structure, mechanism of action and applicability of secreted antimicrobial peptides/proteins
- Antifungal therapeutics and resistance
Epigenetics and Epitranscriptomics
- RNA methylation and its impact on RNA metabolism
- Chromatin assembly mechanisms: Chromatin remodelers and histone variants
General Facts
The Institute of Molecular Biology is home to five independent research groups. Their scientific interests range from diverse aspects of filamentous fungal physiology and metabolism, antifungal and antimicrobial peptides, to the nature and significance of chromatin remodelling and epitranscriptomic mechanisms. The ultimate aim of all research groups is to translate basic scientific discoveries into the diagnosis and treatment of human disease. Continuous third-party funding (such as FWF) of all research groups and the participation of members of the institute in various intra and extramural network activities, such as the FWF-funded PhD programmes “HOROS” and “CBD”, the Euregio programme “SupErA”, the SFB “RNA Deco”, the MUI-funded MYCOS, the European Cooperation in Science and Technology COST (Netskinmodels, FeSImmChemNet) and the Acceleration Program Seed4Innovation of the University of Milan (Italy) are clear evidence of the high standard of research. Staff members at the Institute of Molecular Biology make a significant contribution to curricular teaching and administration activities at MUI. Gerald Brosch is in charge of organising courses and student affairs for the Molecular Medicine Bachelor and Master studies. Beyond this, all group leaders and scientific staff teach lectures, seminars and practical courses as part of the human medicine, molecular medicine (bachelors’ and masters’) and PhD curricula.
Research
Iron Metabolism of Filamentous Fungi: Links to Human Disease
Hubertus Haas
Aspergillus fumigatus is a typical saprobic mould fungus, but also the most common human airborne fungal pathogen in Europe. Owing to unsatisfactory diagnostic and therapeutic options, it causes allergic and life-threatening invasive diseases, depending on the immune status of the patient. Our research goal is the characterisation of fungal metabolism, in order to improve antifungal therapy, the diagnosis of fungal infections and the biotechnological potential of filamentous fungi. The present research focuses on iron homeostasis-maintaining mechanisms of Aspergilli. We have demonstrated that siderophore-mediated iron acquisition and the iron-sensing transcription factor HapX are vital for A. fumigatus’ virulence in murine aspergillosis models. Consequently, fungal iron metabolism is an attractive target for improving antifungal therapy and diagnosing fungal infections.
Major Achievements
Characterisation of siderophore-mediated iron acquisition and storage
Characterisation of fungal mechanisms for iron sensing and regulation
Metabolic linking of iron regulation, e.g. to ergosterol biosynthesis, hypoxia adaptation and leucine biosynthesis
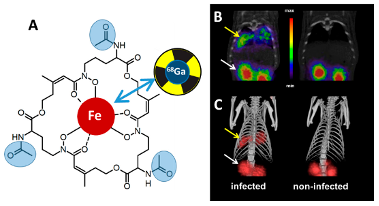
In vivo PET (positron emission tomography) imaging of fungal infections using 68Gallium-labelled siderophores in animal models (Fig. 1, collaboration with Dr. Clemens Decristoforo, Department of Nuclear Medicine)
Proof of concept for siderophores (triacetylfusarinine C) as urine biomarkers for aspergillosis
Future Goals
Further characterisation of fungal iron homeostasis and medical translation. Role of iron and siderophores in microbial interactions.
Functions of Histone-Modifying Enzymes in Gene Regulation and Fungal Physiology
Gerald Brosch, Stefan Graessle and Ingo Bauer
Distinct regulatory sequences in gene promoters are key determinants of gene expression. However, in eukaryotes, the read-out of genetic information is also significantly regulated at the chromatin level. Covalent post-translational modifications (PTMs) of histones and other proteins have profound structural and functional consequences for the transcriptional programme of a cell. Our research focuses squarely on the functional impact of lysine acetylation and arginine methylation on fungal physiology. We are particularly interested in studying the extent of the involvement of modifying activities in fungal pathogenicity and their role in regulating secondary metabolite (SM) production. Our group has generated Aspergillus strains with individual or multiple deletions of all protein arginine methyltransferase (PRMT) genes and all classical lysine deacetylase (KDAC) genes. Using these tools together with specifically engineered transgenes to study the impact on the viability, pathogenicity and metabolism of Aspergilli. Furthermore, we employ proteomic and transcriptomic analyses (i) to characterise novel substrates and (ii) to investigate the contribution of KDACs and PRMTs to the regulation of genes important for fungal virulence and SM production.
Major Achievements
KDACs: For the last ten years, we have characterised KDACs and KDAC complexes in filamentous fungi. We have focused especially on Aspergillus nidulans and its pathogenic relative Aspergillus fumigatus. Members of the genus Aspergillus are important medical pathogens and producers of allergens and highly toxic metabolites such as aflatoxins. Other Aspergilli produce pharmaceutical secondary metabolites (SMs). These include antibiotics and cholesterol-lowering drugs. Interestingly, KDACs play a prominent regulatory role in all these issues.
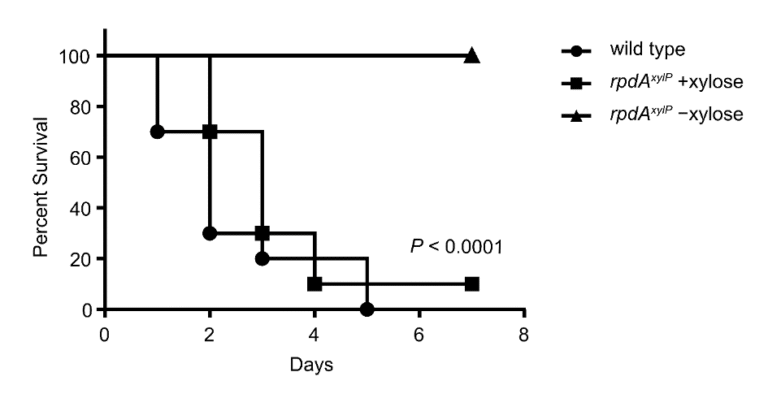
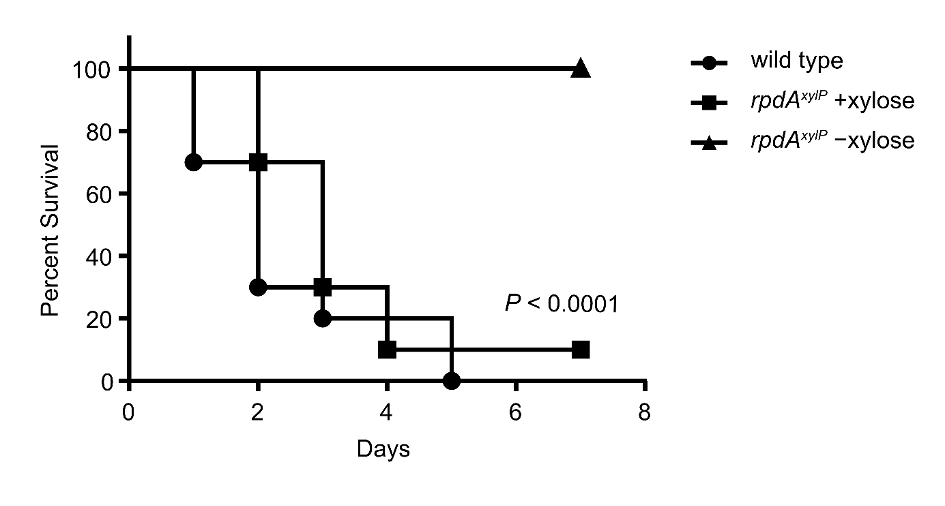
We have identified and functionally characterised two fungus-specific protein domains in the KDAC RpdA. These are essential for the viability of Aspergillus in vitro and may therefore serve as antifungal targets for novel antifungal therapies. We have confirmed that RpdA is an essential fungal virulence factor in vivo in a mouse model for invasive pulmonary aspergillosis (Fig. 2). We have also identified a novel fungus-specific RpdA complex that is essential for the fungus’ sexual reproduction. Another KDAC, HosA, was found to be a major regulator in the biosynthesis of medically important fungal secondary metabolites (e.g. toxins and antibiotics).
PRMTs: Role of protein arginine methylation in stress signalling. Functional linking of Aspergillus type I and II PRMTs to various cellular processes, including detoxification, secondary metabolism, transport and protein secretion. Identification of novel PRMT substrates involved in RNA metabolism.
Future Goals
KDACs: Using a versatile promoter system, various knockout strategies, and transcriptional analyses, we aim to uncover fungal-specific features of RpdA and HosA, identify targets involved in virulence, resistance, and secondary metabolite production, and characterize additional KDAC complexes. We are also investigating the antifungal efficacy of novel synthetic KDAC inhibitors both in vitro and in vivo using different virulence models.
PRMTs: Investigation of the (methyl-) proteome network
Identification and validation of reactive oxygen species (ROS) induced PRMT substrates
Functional consequences of demethylation of ROS-specific substrates
Elucidation of the biological role of as yet uncharacterised type IV PRMTs
Structure, Mechanism of Action and Applicability of Small, Cationic Antimicrobial Peptides and Proteins
Florentine Marx-Ladurner
The rise in drug resistance of human pathogenic bacteria and fungi means we urgently need to search for alternative antimicrobial agents with novel modes of action that differ from existing ones and hamper the onset of resistance development. Promising candidates are small, cationic antimicrobial proteins and peptides (scAMPPs). These are natural defence molecules produced from organisms of all phyla. These scAMPPs are powerful model molecules. They will lead to the development of new effective antimicrobial strategies for therapeutic application.
The Marx group’s mission is clear: to identify and isolate scAMPPs and characterise in detail their mode of action and structure-function relation. The rational design and chemical synthesis of protein and peptide analogues derived from their respective natural parent molecules is the key to vast field of possibilities to improve the antimicrobial efficacy and tolerance by the host and lower the risk for resistance development of pathogenic agents. We are proud to support the Animal Free Research Cluster of the Medical University of Innsbruck. We aim to reduce, refine and replace animal models for early drug screening. We screen scAMPPs in vitro for a selection of the most promising candidates before testing them for their curative potential using two models. The first is an in vitro 3D human reconstructed skin tissue model. The second is an in vivo invertebrate mini-host model infected with opportunistic human pathogens. Both models are advantageous and valid alternatives to in vivo vertebrate models. This approach helps to bridge the gap between in vitro and in vivo drug testing, reduces the number of vertebrate hosts sacrificed for drug screening purposes and lowers the experimental costs.
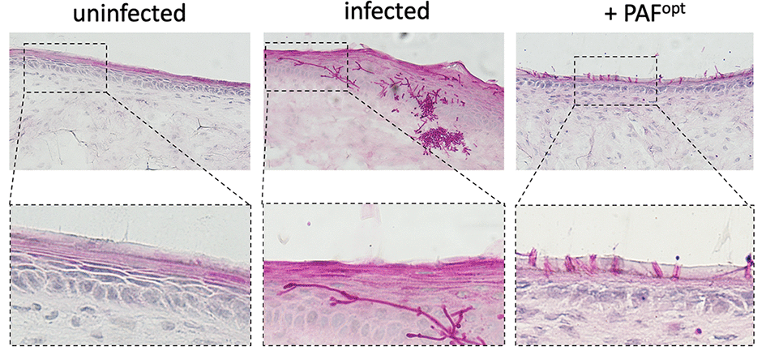
Major Achievements
Understanding the structure-function relationship of scAMPPs and the first steps towards their therapeutic application.
Future Goals
Our aims are:
the rational design to improve the antimicrobial efficacy and species-specificity of scAMPPs and reduce the risk of resistance development in pathogenic microorganisms;
the determination of the mode of action and the identification of molecular targets of scAMPPs;
the combination of scAMPPs with smart drug delivery systems to improve the antimicrobial potential of peptide-based drugs.
Antifungal Therapeutics and Resistance
Fabio Gsaller
Fungi belong to the domain of eukaryotes so that many vital processes in mammalian cells are conserved in fungal species. Substances with antifungal activity often have toxic effects on human cells, which makes the development of new antifungal drugs particularly challenging. In the clinical setting, azole-based antifungals are the first-line treatment of choice for Aspergillus infections. However, emerging resistance to this drug class is a growing threat to public health.
Our research explores the metabolic effects of antifungal drugs and the molecular factors employed by Aspergillus fumigatus to protect itself from the action of anti-mycotic substances. Our work focuses on determining the mode of action and investigating the gene regulatory mechanisms that drive resistance to clinically administered anti-mycotics. This is critical for developing alternative strategies to treat fungal infections.
Major Achievements
Elucidation of the molecular mechanism underlying one of the predominant resistance mechanisms found in A. fumigatus clinical isolates that leads to treatment failure;
Establishment of advanced tools to bolster genetic engineering of Aspergillus fumigatus
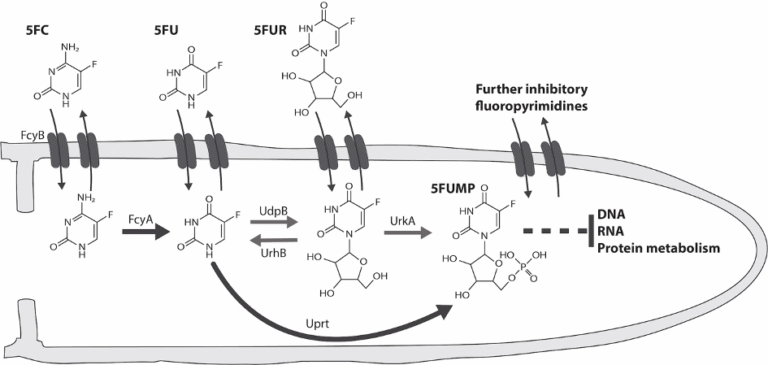
Discovery of major mechanisms that confer intrinsic resistance to the antifungal nucleobase analogue 5-fluorocytosine (Fig. 4).
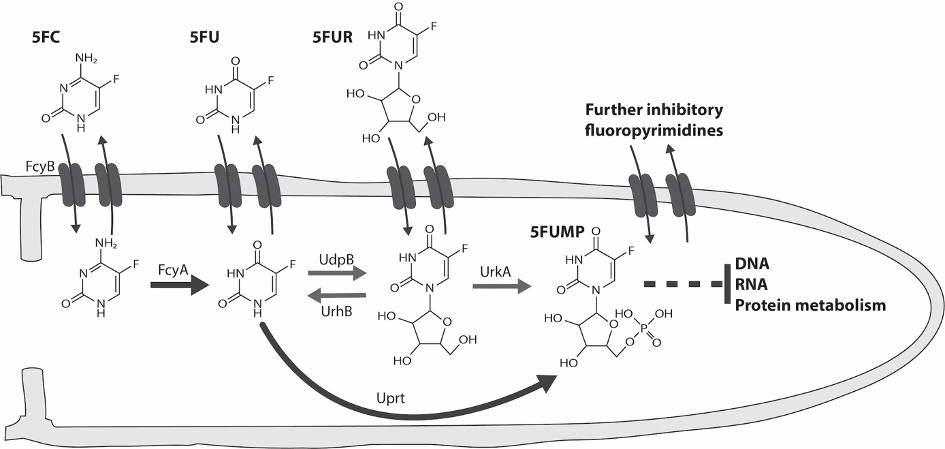
Future Goals
We aim to increase our knowledge of fungal pathogen strategies to counteract the activity of antifungal drugs with the ultimate goal of improving current approaches for the treatment of fungal infections.
Epigenetics and Epitranscriptomics
Alexandra Lusser
Methylation of 5-methylcytosine (m5C) is a prominent and well-characterized modification in tRNAs and rRNAs. However, for mRNAs and other less abundant RNA types, the prevalence and functional significance of m5C remain unclear. Studies conducted across different organisms and tissues have reported widely varying numbers of m5C sites, ranging from several thousand to just a few hundred. In the context of mRNA, m5C plays diverse roles that are still being explored. These include promoting nuclear export by the export factor ALYREF, stimulating or inhibiting translation and increasing mRNA stability via the binding of specific reader proteins. However, it is not yet clear whether m5C modification only has context-dependent functions or whether it plays a more general role. The enzymes that catalyse RNA cytosine methylation belong to the NOL1/NOP2/Sun (NSUN) family of methyltransferases. The predominant substrates of these RCMTs are tRNAs and rRNAs. However, recent reports have linked Nsun2 and Nsun6 to the modification of mRNAs. My laboratory is currently researching the following issues: the analysis of 5-methylcytosine distribution and function in mammalian and viral mRNA, the functional analysis of RNA cytosine methyltransferase enzymes, the analysis of RNA turnover dynamics and the development of RNA aptamer-based tools for RNA imaging and enrichment.
Major Achievements
The mapping of m5C sites in mRNA remains a significant challenge in the field. In the past report period, we improved the bisulfite-sequencing technique and the data analysis pipeline. We did this by including additional filters that were tailored towards the elimination of false positive m5C calls. We applied the updated experimental BS-seq conditions to analyse m5C modification of SARS-CoV-2 RNA. Contrary to previous reports, our findings revealed no substantial m5C modification in SARS-CoV-2 RNA isolated from various cell types. However, we did identify the expected m5C sites in host tRNAs and mitochondrial rRNA. We have conclusively demonstrated that previously reported m5C sites in two other RNA viruses (HIV and MMV) cannot be confirmed, regardless of whether direct PCR-based bisulfite analysis or re-analysing the original published data sets using meRanTK v.1.3.0. is employed (Fig. 5). The data strongly support the need for meticulous design and analysis of studies dealing with epitranscriptomic modifications, particularly m5C in RNA, to prevent false positive calls.
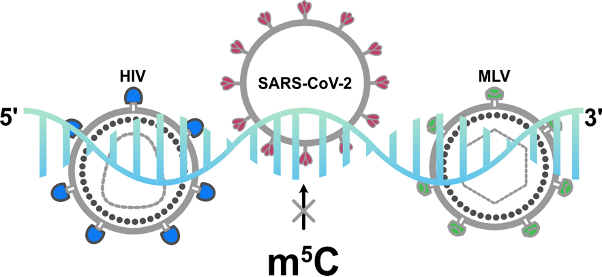
Future Goals
Future research goals include the study of RNA turnover dynamics using the TUC-seq method, which we have previously developed, during embryonic stem cell (ESC) differentiation and examining the influence of RNA cytosine methyltransferases on these processes. Additionally, we aim to investigate the potential of employing RNA aptamers for RNA imaging within cells, which could provide valuable insights into RNA behaviour and function in various cellular contexts.
Pictures
Selected Publications
Oberegger, Simon; Misslinger, Matthias; Faserl, Klaus; Sarg, Bettina; Farhan, Hesso; Haas, Hubertus: The cytosolic form of dual localized BolA family protein Bol3 is important for adaptation to iron starvation in Aspergillus fumigatus. OPEN BIOL. 2024 14(6):240033.
Yap, Annie; Volz, Ricarda; Paul, Sanjoy, Moye-Rowley, W Scott; Haas, Hubertus: Regulation of high-affinity iron acquisition, including acquisition mediated by the iron permease FtrA, is coordinated by AtrR, SrbA, and SreA in Aspergillus fumigatus. MBIO. 2023;14(3):e0075723.
Bruch, Alexander; Lazarova, Valentina; Berg, Maximilian; Kruger, Thomas; Schauble, Sascha; Kelani, Abdulrahman A; Mertens, Birte; Lehenberger, Pamela; Kniemeyer, Olaf; Kaiser, Stefanie; Panagiotou, Gianni; Gsaller, Fabio; Blango, Matthew G.: tRNA hypomodification facilitates 5-fluorocytosine resistance via cross-pathway control system activation in Aspergillus fumigatus. NUCLEIC ACIDS RESEARCH. 2024; 53(3);
Storer, Isabelle; Sastre-Velasquez, L.E.; Easter, T.; Mertens, B.; Dallemulle, A.; Bottery, M.; Tank, R.; Offterdinger, M.; Bromley, M.J.; van Rhijn, N.; Gsaller, F.: Shining a light on the impact of antifungals on Aspergillus fumigatus subcellular dynamics through fluorescence imaging. ANTIMICROBIAL AGENTS AND CHEMOTHERAPY. 2024; 68(11);
Huang, Anming; Riepler, Lydia; Rieder, Dietmar; Kimpel, Janine; Lusser, Alexandra: No evidence for epitranscriptomic m5C modification of SARS-CoV-2, HIV and MLV viral RNA.
RNA. 2023; 29(6); 756-763.
Berg, Kevin; Lodha, Manivel; Delazer, Isabel; Bartosik, Karolina; Garcia, Yilliam Cruz; Hennig, Thomas; Wolf, Elmar; Doelken, Lars; Lusser, Alexandra; Prusty, Bhupesh K.; Erhard, Florian: Correcting 4sU induced quantification bias in nucleotide conversion RNA-seq data.
NUCLEIC ACIDS RESEARCH. 2024; 52(7)
Bereiter Raphael; Flemmich, Laurin; Nykiel, Kamila; Heel, Sarah; Geley, Stephan; Hanisch, Malou; Eichler, Clemens; Breuker, Kathrin; Lusser, Alexandra; Micura, Ronald: Engineering covalent small molecule-RNA complexes in living cells. NAT. CHEM. BIOL. 2025; Epub ahead of print.
Akkus-Dagdeviren Zeynep Burcu; Saleh Ahmad; Schöpf Cristina; Truszkowska Martyna; Bratschun-Khan Doris; Fürst Andrea; Seybold Anna; Offterdinger Martin; Marx Florentine; Bernkop-Schnürch A. Phosphatase-degradable nanoparticles: A game-changing approach for the delivery of antifungal proteins. J COLLOID INTERFACE SCI 2023, 646:290-300. doi: 10.1016/j.jcis.2023.05.051.
Váradi Györgyi; Batta Gyula: Galgóczy László; Hajdu Dorottya; Fizil Ádam; Czajlik András; Virágh Máté; Kele Zoltán; Meyer Vera; Jung Sascha; Marx Florentine; Tóth Gábor K. Confirmation of the disulfide connectivity and strategies for chemical synthesis of the four-disulfide-bond-stabilized Aspergillus giganteus antifungal protein, AFP. J NAT PROD 2023, 86:782-790. doi:10.1021/acs.jnatprod.2c00954.
Karahoda, Betim; Pardeshi, Lakhansing; Ulas, Mevlut; Dong, Zhiqiang; Shirgaonkar, Niranjan; Guo, Shuhui; Wang, Fang; Tan, Kaeling; Sarikaya-Bayram, Özlem; Bauer, Ingo; Dowling, Paul; Fleming, Alastair B; Pfannenstiel, Brandon T; Luciano-Rosario, Dianiris; Berger, Harald; Graessle, Stefan; Alhussain, Mohamed M; Strauss, Joseph; Keller, Nancy P; Wong, Koon Ho; Bayram, Özgür: The KdmB-EcoA-RpdA-SntB chromatin complex binds regulatory genes and coordinates fungal development with mycotoxin synthesis. NUCLEIC ACIDS RESEARCH. 2022; 50(17); 9797-9813.
Bauer, Ingo; Graessle, Stefan: Fungal Lysine Deacetylases in Virulence, Resistance, and Production of Small Bioactive Compounds. GENE. 2021; 12(10); 1470.
Selection of Funding
P 35951 Stand-Alone Project Austrian Science Fund (FWF) to FG
FWF SFB F8009-B to AL
MUI MYCOS to FG
FWF CBD to HH
Collaborations
- Laura Alcazar-Fuoli, Instituto de Salud Carlos III, Madrid, Spain
- Andreas Bernkop-Schnürch, University of Innsbruck, Innsbruck, Austria
- Axel Brakhage, F. Schiller University Jena, Germany
- Franz Bracher, LMU Munich, Munich, Germany
- Mike Bromley, The University of Manchester, UK
- Matthew Blango, Leibniz Institute for Natural Product Research and Infection Biology-Hans Knöll Institute (Leibniz-HKI), Germany
- Ronald Micura, University of Innsbruck, Austria
- Kathrin Breuker, University of Innsbruck, Austria
- Florian Erhard, Julius-Maximilians-Universität Würzburg, Germany
- László Galgóczi, University of Szeged, Szeged, Hungary
- Ilse Jacobsen, Hans-Knöll Institut, Jena, Germany
- Nermina Malanovic, University of Graz, Graz, Austria
- Nir Osherov, Tel-Aviv University, Israel
- Alessandra Romanelli, University of Milan, Milan, Italy
- Özgür Bayram, Maynooth University, Maynooth, Ireland
- Joseph Strauss, BOKU, Vienna, Austria
- Györgyi Váradi, University of Szeged Albert Szent-Györgyi Medical School, Szeged, Hungary