Schöpfstrasse 41
6020 Innsbruck, Austria
Email: florian.kronenberg@i-med.ac.at
Website: https://genepi.i-med.ac.at/
Research Branch (ÖSTAT Classification)
102004,102010, 301301, 301302, 303007
Keywords
cardiovascular disease, complex phenotypes, computational genetics, Genetic epidemiology, genome-wide association studies, genotype imputation, kidney disease, lipoprotein metabolism, mitochondrial DNA, and risk factors
Research Focus
We aim to identify determinants of health and disease-related to genetic variability, environmental components and biochemical parameters and study their physiological or pathophysiological functions.
Our phenotypes of interest are complex in nature due to an interplay of these factors and are related to atherosclerosis, diabetes mellitus, metabolic syndrome, cancer, infertility, neurodegeneration and associated intermediate phenotypes such as lipoprotein metabolism and inflammation.
Our Institute serves as a bridge between basic and clinical research. We work with genetic-epidemiological methods with association studies of candidate genes as well as hypothesis-free genome-wide association studies and clinical-epidemiological methods. For functional studies, we use a variety of cell culture models implementing state-of-the-art methods of protein chemistry and molecular biology. Finally, we develop bioinformatics solutions for problems related to medical genetic research and diagnosis.
General Facts
The institute, which has about 20 members (Figure 1), serves as a bridge between basic and clinical research. We have three legs, the strength of which lies in interconnected collaboration: 1) a protein chemistry and cell culture laboratory undertakes a variety of structural-functional and epidemiological studies into various phenotypes related to lipoprotein metabolism and other metabolic phenotypes (e.g. Lp(a), apolipoprotein A-IV, afamin, PCSK9, cholesterol efflux assays); 2) a molecular genetics laboratory performs sequencing and genotyping for various projects, with a strong focus on mitochondrial DNA as well as complex genetic regions such as the K-IV type 2 region of the LPA gene – new technologies such as nanopore sequencing have recently become a major focus of this research; 3) the computational and statistical genetics laboratory focusses on statistics, epidemiology, computer science and bioinformatics, and represents an important link between the various research groups. A close interconnection of these three “units” to form a strongly collaborative alliance has become increasingly evident in recent years. There are barely any major projects that involve fewer than two of these main groups. The output and success of the institute are based upon constant dialogue between the various disciplines in a problem-oriented and critical elucidation of research questions.
Research
Lipoprotein(a)
Stefan Coassin, Sebastian Schönherr, Claudia Lamina, Florian Kronenberg
High concentrations of lipoprotein(a) [Lp(a)] are a strong risk factor for cardiovascular disease and are mostly genetically influenced by the LPA gene. Despite this strong genetic regulation, Lp(a) concentrations in individuals with the same isoform combination can vary 200-fold. This suggests that Lp(a) levels are modified by additional genetic variants, which have to be identified. However, the LPA gene contains a large, hypervariable region, which is not properly covered by any genome reference database. This region referred to as “KIV-2 repeat” can encompass up to 70% of the gene and may therefore contain most of the genetic variability in LPA. We pursue the following research aims.
1. Identification of new genetic variants regulating Lp(a)
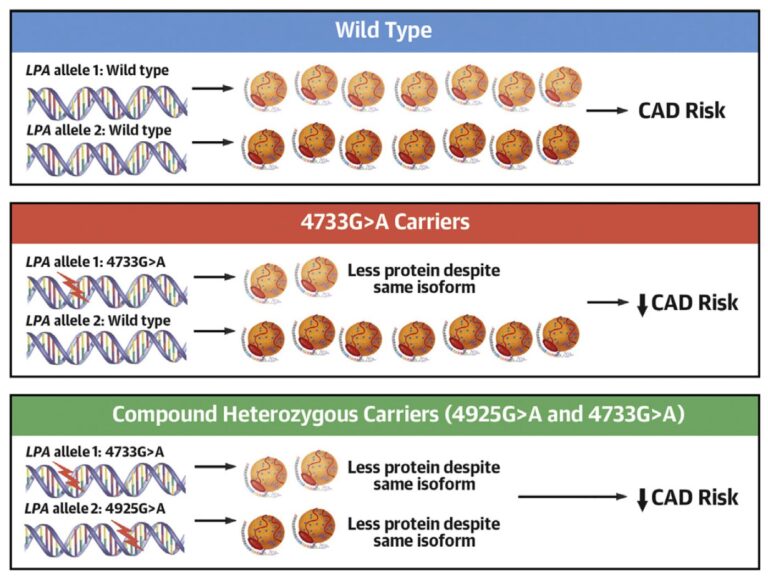
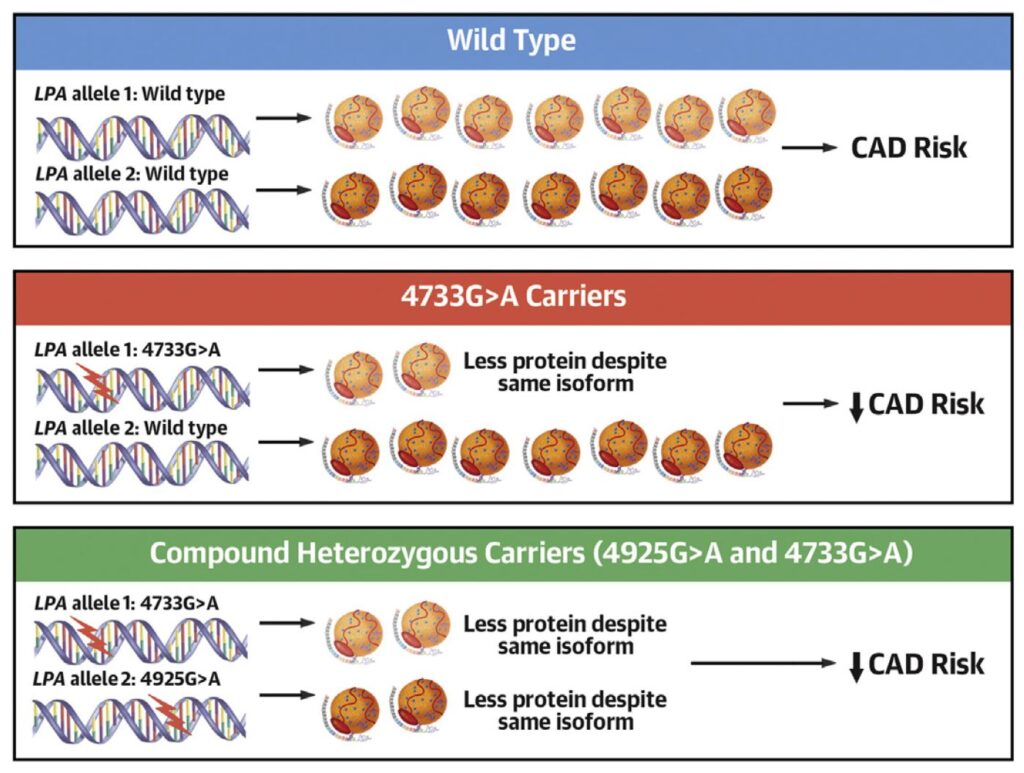
A major part of our work concerns the investigation of mutations in the KIV-2 repeat region of the LPA gene and their interaction with SNPs in the non-repetitive region of LPA. By combining the expertise of the institute in the detection of low-level mutations and our long-standing experience in Lp(a) research, we identified a mutation hidden in the non-coding part of the KIV-2 region that modifies LPA mRNA splicing. After developing a tailored high-throughput assay, we screened >4,600 German individuals and found that this mutation is extremely frequent (~38% of all individuals) and generates a lower-than-expected Lp(a) concentration in these individuals, providing a molecular explanation for the enormous variance of Lp(a) concentrations in the population. We demonstrated that this mutation lowers cardiovascular risk significantly and is indeed the most important modifier of Lp(a) concentrations in the general European population (Figure 2) – yet, it was hidden in plain sight by its location in the complex KIV-2 region (J Am Coll Cardiol 2021), until now. Moreover, in two further studies we identified the genetic mechanisms that form the basis of the Lp(a)-lowering effect of two LPA mutations that have been known for ~25 years, whereas its Lp(a)-lowering effect could not be explained so far (Atherosclerosis 2022a; J Lipid Res 2022). Finally, we developed a general approach capable of tackling variants in complex genome regions in short-read sequencing data from public biobanks such as the UK Biobank. This resource contains genetic and phenotypical data from up to 500,000 individuals across the United Kingdom. Currently, we are using 200,000 exome sequencing datasets to search for new genetic variants in the KIV-2 region and to search for novel regulators of Lp(a) concentrations.
2. Lp(a) and SARS-CoV-2 infections
Due to the high homology of apo(a) with plasminogen, Lp(a) has been repeatedly suggested as a risk factor for thromboembolic events. As patients with SARS-CoV-2 infections have a markedly increased risk for thromboembolic events, we evaluated whether or not SARS-CoV-2 infections modify the risk of high Lp(a) concentrations for ischemic heart disease or thromboembolic events during the first 8.5 months follow-up of the pandemic. We used data from the UK Biobank during the first 8.5 months of the SARS-CoV-2 pandemic. The risk for ischemic heart disease increased with higher Lp(a) concentrations in both, the 435,104 population controls and the 6,937 SARS-CoV-2 positive patients. The causality of the findings was supported by a genetic risk score for Lp(a). A SARS-CoV-2 infection modified the association with a steeper increase in risk for infected patients (interaction p-value = 0.03). Most importantly, although SARS-CoV-2 positive patients had a five times higher frequency of thromboembolic events compared to the population controls (1.53% vs. 0.31%), the risk was not influenced by Lp(a). In summary, SARS-CoV-2 infections enforce the association between high Lp(a) and ischemic heart disease but the risk for thromboembolic events is not influenced by Lp(a) (J. Intern. Med. 2022a).
3. Special Issue on Lp(a) published in Atherosclerosis
Marlys L. Koschinsky from the University of Western Ontario, Canada, and Florian Kronenberg organized and edited a special issue on the topic Lp(a) with 13 review articles and further 15 original papers on Lp(a) which have been printed in the May 2022 issue of “Atherosclerosis” (Atherosclerosis 2022c). Five of the 13 reviews were written by coworkers from the institute together with external experts. This special issue can be considered as the most up-to-date overview on Lp(a).
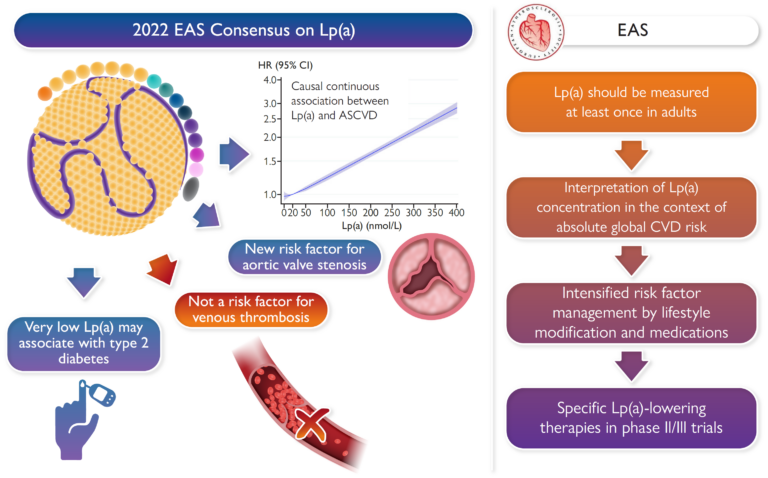
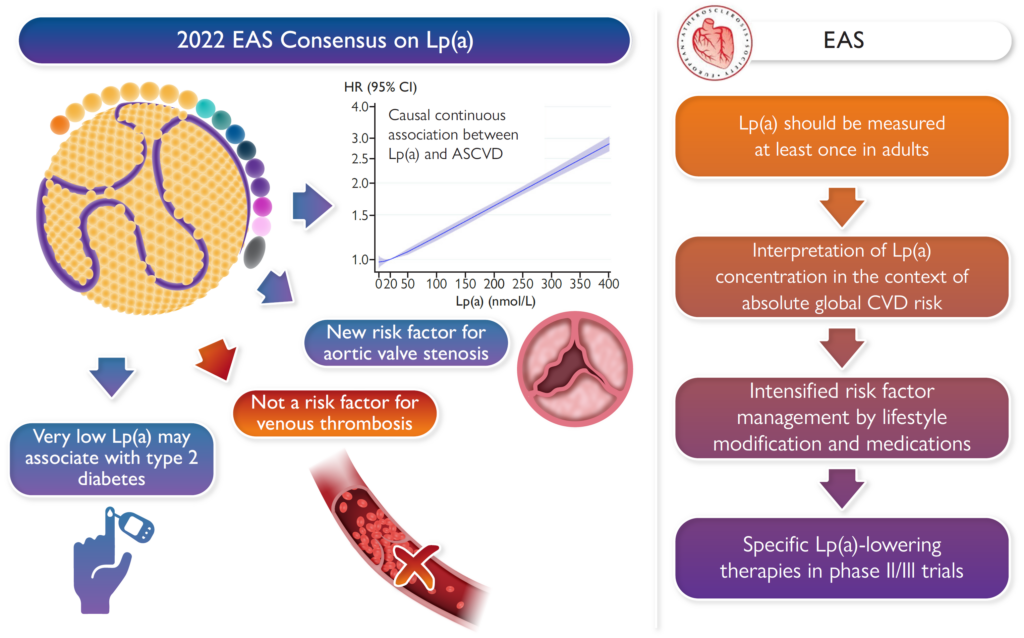
Lp(a) Consensus paper of the European Atherosclerosis Society (EAS)
Florian Kronenberg
Together with further 19 Lp(a) experts from all over the world, the co-chairs Florian Kronenberg, Samia Mora and Erik Stroes provided an overview on the latest developments and recommendations on Lp(a). Based on epidemiological and genetic findings, it is recommended that Lp(a) should be measured at least once in all adults to identify those with high cardiovascular risk. It is described how Lp(a) can be incorporate in clinical risk estimation, and what to do in case of high Lp(a) (Eur. Heart J. 2022). The following graphical abstract describes the main points of the Lp(a) consensus paper.
Biomarkers
Claudia Lamina, Barbara Kollerits, Johanna Schachtl-Rieß, Florian Kronenberg
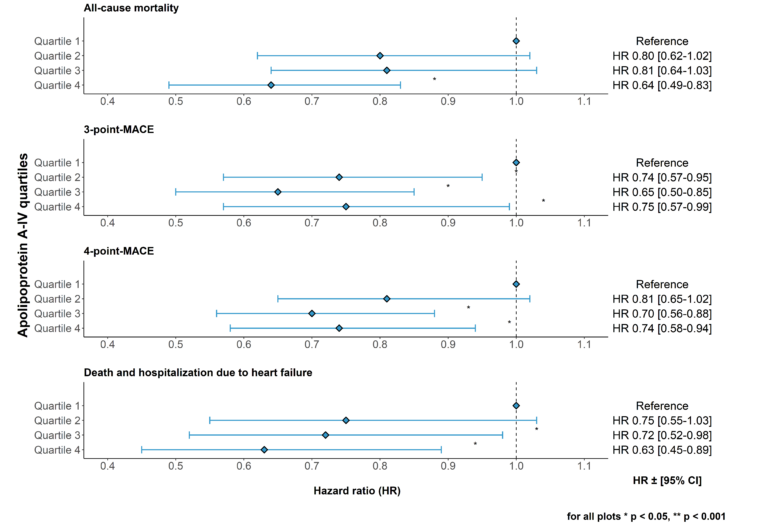
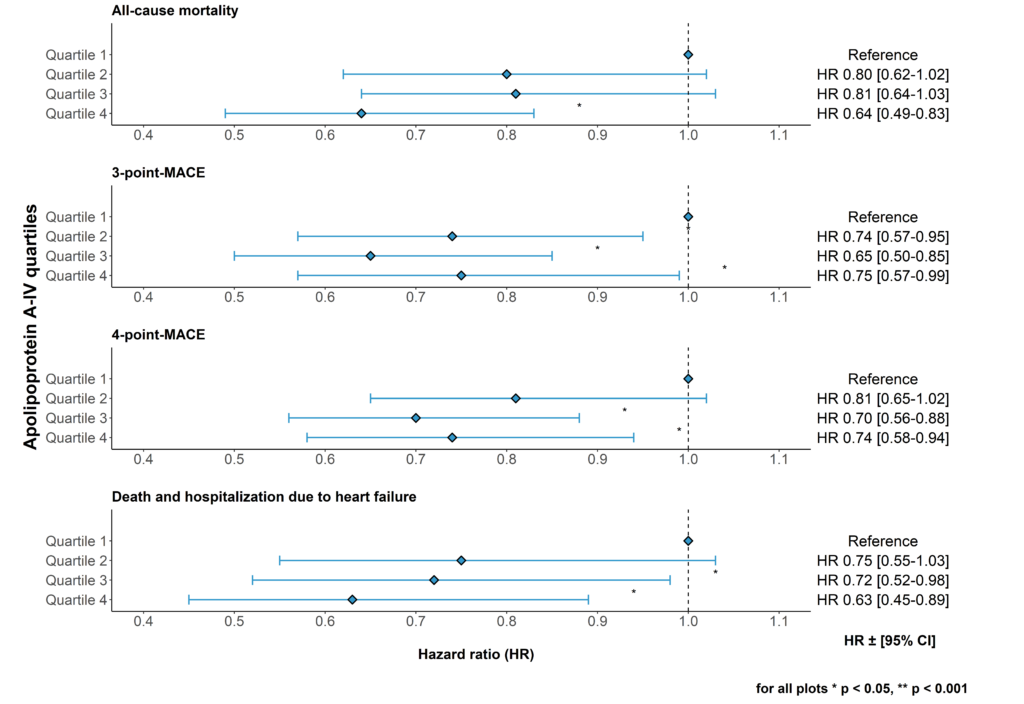
1. Apolipoprotein A-IV and risk for cardiovascular outcomes and mortality in chronic kidney disease
Apolipoprotein A-IV (apoA-IV) has antiatherogenic, anti-oxidative, anti-inflammatory and anti-thrombotic properties, and levels increase significantly during the course of chronic kidney disease. We recently investigated the association between apoA-IV and all-cause mortality and cardiovascular outcomes in more than 5000 patients of the German Chronic Kidney Disease (GCKD) study. We found that apoA-IV is an independent risk marker for reduced all-cause mortality, cardiovascular events and heart failure in a large cohort of patients with chronic kidney disease (Figure 4) (J. Intern. Med. 2022b).
2. Cholesterol efflux capacity
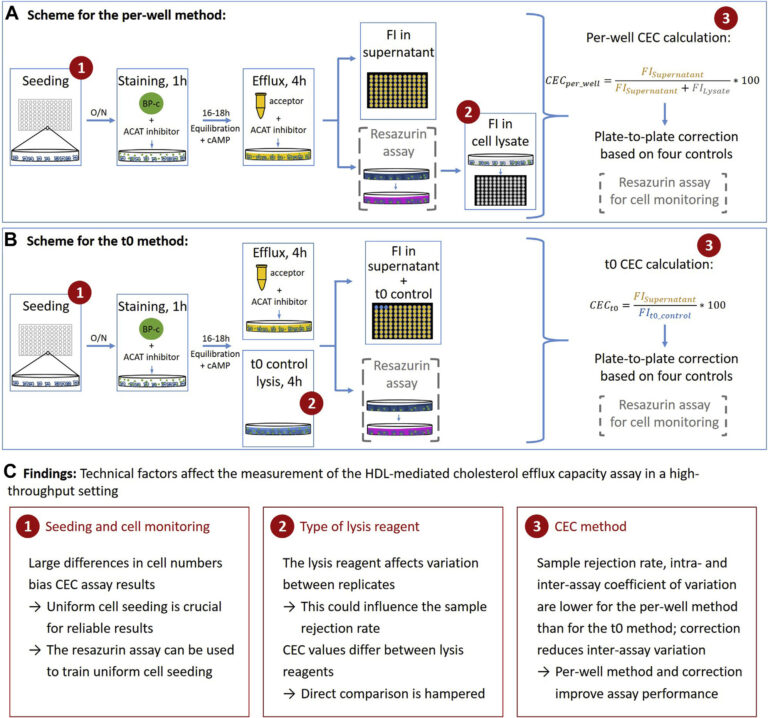
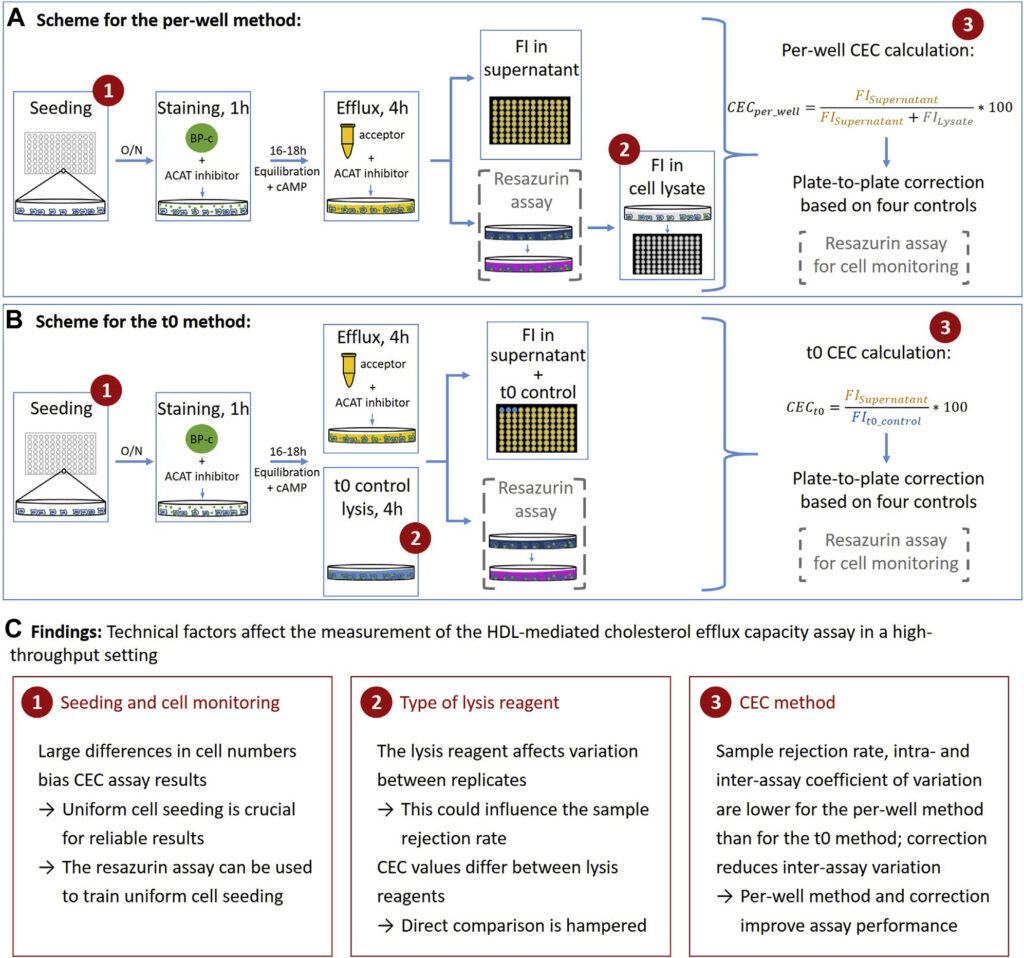
HDL-mediated cholesterol efflux capacity (CEC) is defined as the ability of HDL particles to induce efflux of cholesterol from peripheral cells such as macrophages. It can be measured in vitro using a cell-based assay. Epidemiological studies have shown that it is inversely associated with cardiovascular disease in the general population and may protect from atherosclerosis; however, many open questions remain.
We are interested in identifying genetic and non-genetic determinants of CEC and understanding whether CEC is also associated with diseases in other cohorts. Since CEC measurements in large study cohorts are required to address these questions, but the assay is not well standardized and different protocols have been published, we are also interested in the technical aspects of CEC measuring. To develop a standardized high-throughput protocol, we 1) searched published CEC protocols for technical differences, 2) systematically tested technical factors that can influence CEC measurement and 3) tested a high-throughput CEC protocol and quality control measures to determine a reliable high-throughput CEC protocol. We identified the type of lysis reagent, cell numbers and calculation method of CEC as technical factors that influence the quality of CEC measurements (Figure 5) and provide a detailed protocol with extensive quality control to measure CEC (J Lipid Res 2021). We are currently using this CEC protocol to measure CEC in large cohorts and to investigate genetic and non-genetic determinants of CEC.
3. PCSK9 and cardiovascular disease in patients with moderately decreased kidney function
Only few and mostly small studies with contradicting results have investigated the association between PCSK9 serum concentrations and cardiovascular outcomes in chronic kidney disease (CKD) patients. In a prospective study with more than 5,000 high-risk patients with moderately severe CKD from the German Chronic Kidney Disease study over a period of 6.5 years, we observed that higher baseline serum PCSK9 concentrations are associated with prevalent and incident cardiovascular disease independent of major confounders. This observation argues for a pronounced lipid-lowering therapy especially in patients with prevalent CVD (Clin. J. Am. Soc. Nephrol. 2022).
Mitochondrial DNA
Hansi Weissensteiner, Lukas Forer, Florian Kronenberg, Sebastian Schönherr
The group has a strong interest in the analysis of the mitochondrial genomes (mtDNA) and related research. The focus of the research throughout the past two years 2021-22 was primarily on benchmarking mtDNA analysis from different novel sequencing approaches, establishing methods for the analysis and quality control of mtDNA, and genetic population studies based on the maternally inherited mitochondrial genome. Furthermore, we investigated the association of mtDNA copy numbers with metabolic syndrome and diabetes mellitus.
1. Benchmarking mtDNA variant calling
With the appearance of novel sequencing protocols / devices / methods, it is crucial to assess the accuracy of the newly generated data. As there are hundreds to thousands of mitochondrial genomes per cell, we are often interested in the homogeneity / heterogeneity of mitochondrial genomes. Here, the term heteroplasmy describes that differing mitochondrial genomes are within a sample, which can often have a functional role, especially in cancer genetics or ageing related processes. Therefore, accurate analysis of heteroplasmy is crucial and benchmarking new methods allows identifying strength and limitations. While we benchmarked so called Next-Generation Sequencing approaches (Int. J. Mol. Sci. 2021a, Int. J. Mol. Sci. 2021b) – we also assessed the latest methods, so called Third-Generation Sequencing or long-read sequencing approaches for estimating the detection limits (Front. Genet. 2022).
2. Quality Control of mtDNA
Early 2021 we published Haplocheck, a tool for quality control not only in mtDNA sequencing studies. The software allows the fast detection of contamination in large NGS sequencing projects by inspecting polymorphic sites in the mitochondrial genome (Genome Res. 2021). As the mitochondria content surpasses nuclear DNA content often by a factor of = 100 or more, its size is only a fraction so that the method is a quick proxy for contamination. We further integrated it into a central web service (https://mitoverse.i-med.ac.at)
3. Maternal genetic history in Cambodia
Our group analyzed the mitochondrial DNA of 299 Cambodian refugees from three different ethnic groups covering most Cambodian provinces. We found that the population was genetically isolated, with no genetic barriers detected between the ethnicities. We observed several autochthonous maternal lineages, indicating the genetic isolation. For this purpose, we also extended our Haplogrep software for the generation of phylogenetic trees. As Cambodia is often underrepresented in sequencing projects, our data is the largest complete sequence mtDNA cohort to date (Sci. Rep. 2021).
4. mtDNA copy number and metabolic syndrome and type 2 diabetes
We investigated the association of mtDNA copy number with type 2 diabetes and the potential mediating role of metabolic syndrome in two independent studies: 4812 patients from the German Chronic Kidney Disease (GCKD) study and 9364 individuals from the Cooperative Health Research in South Tyrol (CHRIS) study (J. Intern. Med. 2021). In both studies, mtDNA copy number showed a significant correlation with most metabolic syndrome parameters: the mtDNA copy number decreased with the increasing number of metabolic syndrome components. Furthermore, individuals with a low mtDNA copy number had significantly higher odds of metabolic syndrome and type 2 diabetes mellitus. A major part of the total effect of mtDNA copy number on type 2 diabetes was mediated by obesity parameters.
Computational Genomics
Sebastian Schönherr, Lukas Forer, Claudia Lamina, Florian Kronenberg, Hansi Weissensteiner
We develop novel algorithms and computational tools to explore the genetics of various phenotypes.
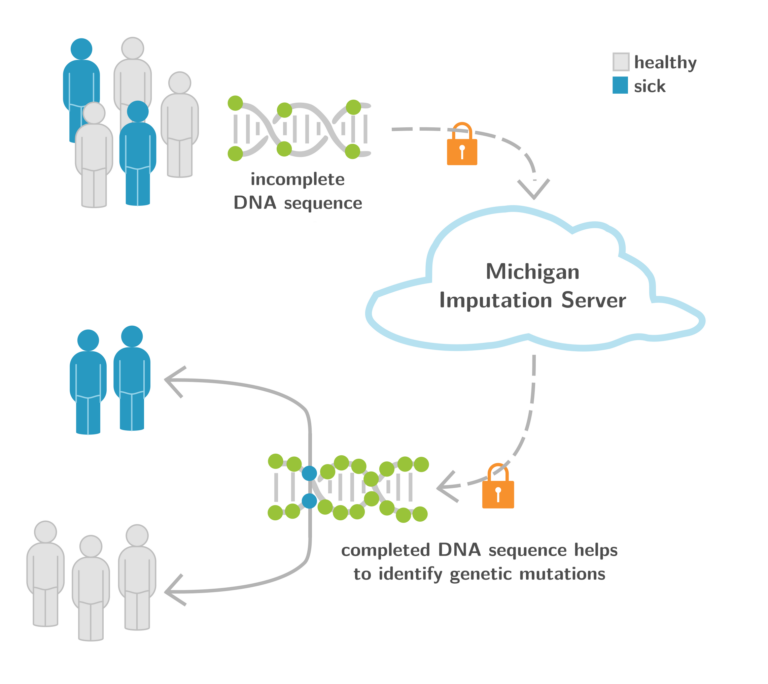
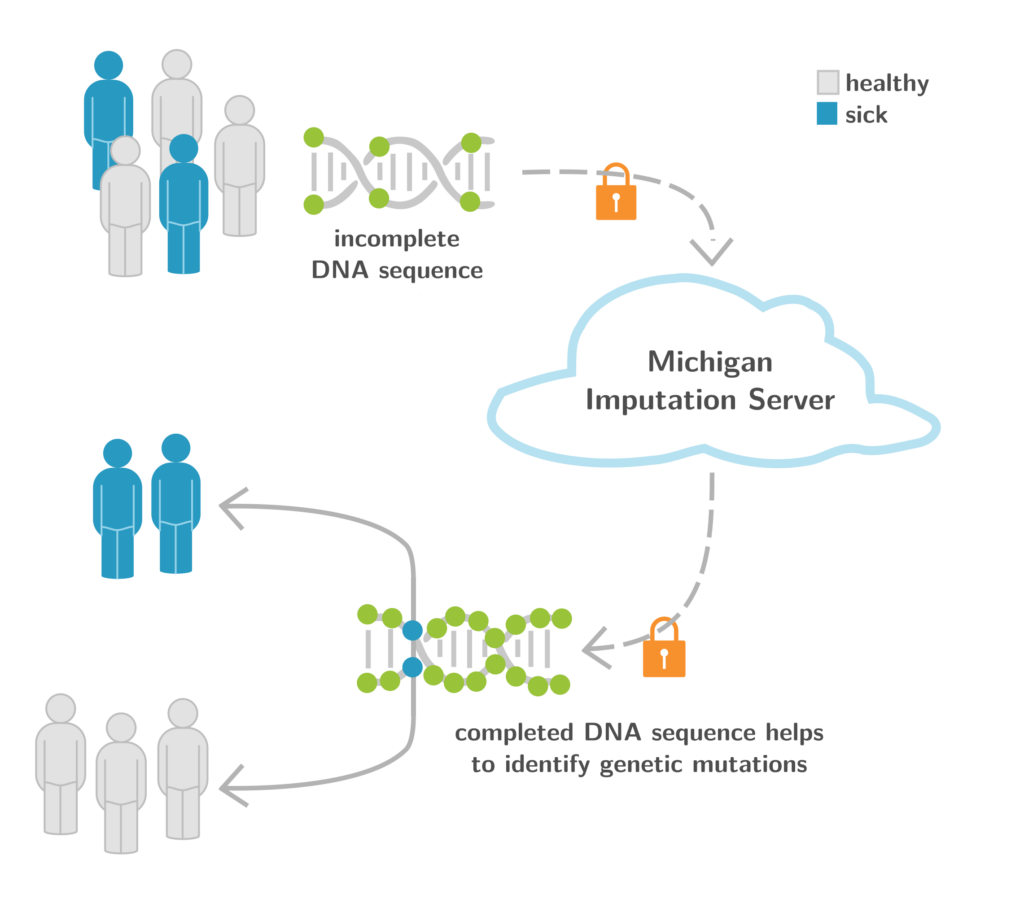
1. Michigan Imputation Server as a Backbone for GWA Studies Worldwide
The Michigan Imputation Server (MIS, https://imputationserver.sph.umich.edu) (Figure 6) is an ongoing collaboration with the University of Michigan (USA) and the EURAC Research (Italy). It provides a free genotype imputation service for Genome-Wide Association Studies (GWAS), a necessary step for each analysis. We are excited to report that we recently crossed the 100 million mark of imputed genomes worldwide. With >10,000 users and >2,400 citations of this Nature Genetics paper published in 2016, MIS is the central backbone for GWAS worldwide. One aim of the Michigan Imputation Server is to provide population-specific reference panels to improve imputation for non-European individuals. For example, the Michigan Imputation Server supports the Genome Asia reference panel, which includes a whole-genome sequencing reference dataset from 1,739 individuals in 219 population groups and 64 countries across Asia, and facilitates genetic studies of Asian populations (Nature 2019). Another example is the TOPMed-based imputation reference panel, which includes 97,256 individuals with 308,107,085 SNPs and Insertion-Deletions (InDels) (TOPMed Imputation Server) (Am. J. Hum. Genet. 2022). We are also providing an HLA reference panel for COVID-19 related studies. All services and reference panels mentioned are free of charge and accessible for academic and non-academic users worldwide.
The current focus of this research project is to extend the functionality and add new tools for the research community. Recently, we added the Rsq-Browser application (https://imputationserver.sph.umich.edu/rsq-browser) that allows searching and comparing imputation qualities among different populations, genotyping arrays, and reference panels.
2. Computation and application of genetic risk scores
With the emerging availability of reasonably priced SNP-chips, polygenic risks scores, which can involve up to several millions of genetic variants, are more frequently applied in epidemiological studies, but are also discussed for clinical applications. To aid researchers in using these scores in their studies, we developed the pgs-calc program, which is available as a web-service on https://imputationserver.sph.umich.edu or on GitHub (https://github.com/lukfor/pgs-calc) for local usage. This broad availability of genetic risk scores for common diseases improves risk prediction: however, it simultaneously raises the question of whether assessing family history might have become redundant. We evaluated the amount of shared information between family history and genetic risk and investigated their combined effect on risk of stroke, myocardial infarction, and type 2 diabetes. We found that both, genetic information and family history are relevant for the prediction of those diseases independently of each other and might be used for identification of high-risk groups to personalize prevention measures (Atherosclerosis 2022b, Stroke 2022).
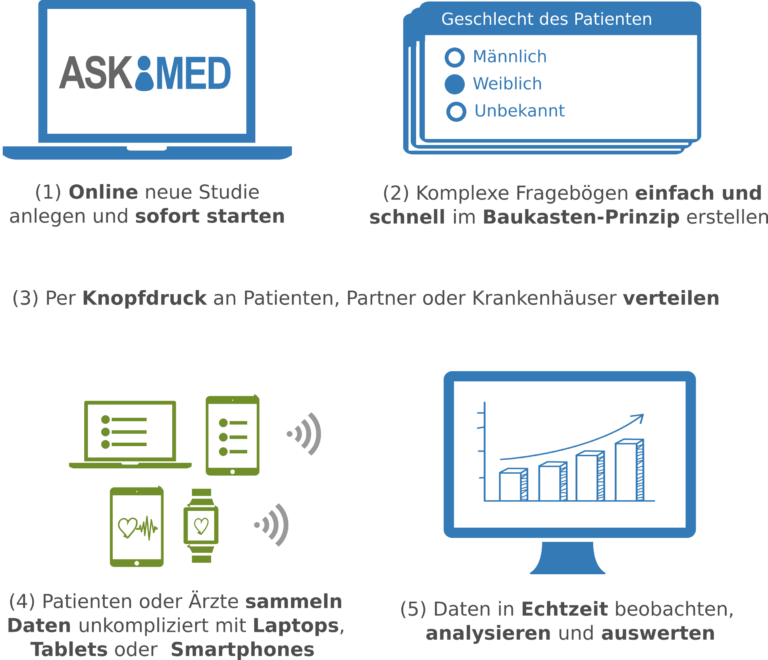
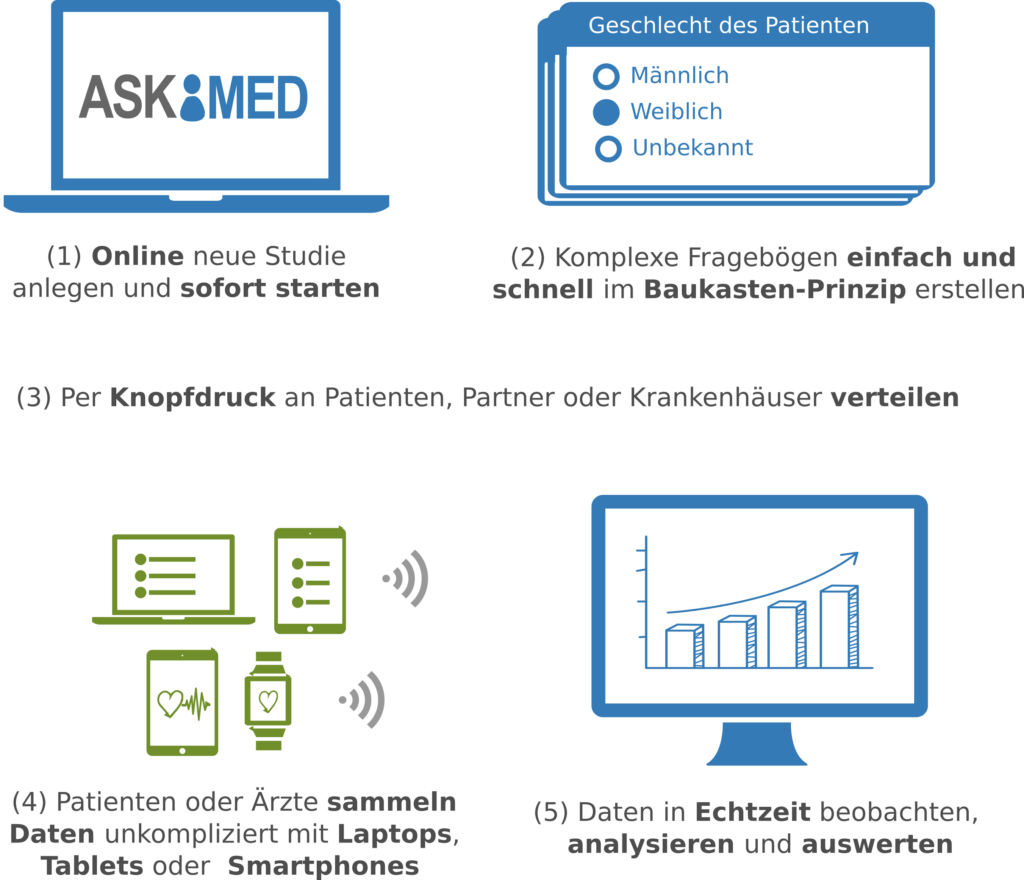
Askimed – a eCRF System for Medical Studies in the Cloud
Lukas Forer, Hansi Weissensteiner, Florian Kronenberg, Sebastian Schönherr
High-quality data collection is key for clinical trials and registries. To collect data in a digital way, we have developed the Askimed eCRF software. Askimed (https://www.askimed.com) described in Figure 7 is a cloud-based web platform that provides an electronic case report forms (eCRF) solution including the following features: (a) data collection based on an CRF, (b) data management including a permission system for the study of collaborators, (c) data preparation for data analysis and (d) survey functionality. Askimed offers a hosted version for our cooperation partners, and we currently support over 15 trials and registries from the Medical University of Innsbruck (e.g. Endometriosis Registry, ACEI-COVID Study, FH Registry, HEVACC-Study, Shieldvacc-Study).
Structural Biology of Therapeutic Protein Targets and Psychotropic Drug Biosynthesis
Sebastiaan Werten, Andreas Naschberger, Bernhard Rupp
The structural biology team supports multiple interdisciplinary projects with detailed 3-dimensional molecular structures of disease-related proteins by means of experimental X-ray diffraction methods, cryo-EM, and in-silico modelling.
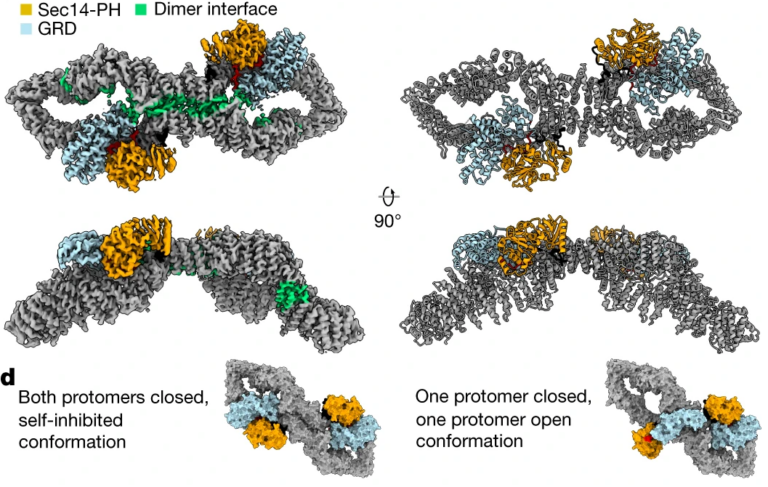
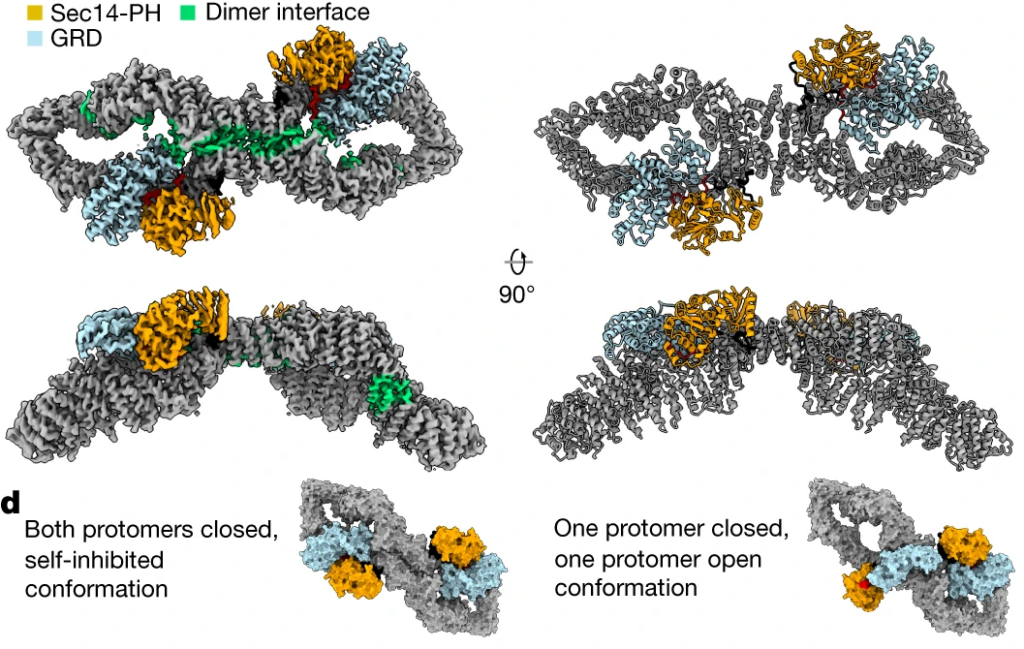
1. The Structure of Neurofibromin.
In collaboration with the Cryo-EM facility in Stockholm, we lead the high-profile publication of the neurofibromin structure in Nature 2021. The autosomal dominant monogenetic disease neurofibromatosis type 1 (NF1) affects approximately one in 3,000 individuals and is caused by mutations in the NF1 tumor suppressor gene, leading to dysfunction in the protein neurofibromin (Nf1). As a GTPase-activating protein, a key function of Nf1 is repression of the Ras oncogene signaling cascade. In the closed conformation, HEAT/ARM core domains shield the GTPase-activating protein-related domain (GRD), so that Ras binding is sterically inhibited (Figure 8). The transition between closed, self-inhibited states of Nf1 and open states provides guidance for targeted studies deciphering the complex molecular mechanism behind the widespread neurofibromatosis syndrome and Nf1 dysfunction in carcinogenesis.
2. Psilocybin Biosynthesis:
As lead investigators of a FWF-funded international DACH project, we examine the biosynthesis of psilocybin, the active psychotropic drug in magic mushrooms by means of X-ray diffraction. Based on in silico modelling and biochemical mutant kinetics analysis we were able to identify psiD as a non-canonical PLP-independent PSD-II decarboxylase, where the aspartate residue of the canonical serin protease catalytic triade DHS is replaced by glutamate; the work has been published jointly by the DACH team (Chembiochem 2022). Work on the experimental verification of the model by a crystal structure is progressing.
Pictures
Selected Publications
Hanks SC, Forer L, Schönherr S, LeFaive J, Martins T, Welch R, Gagliano Taliun SA, Braff D, Johnsen JM, Kenny EE, Konkle BA, Laakso M, Loos RFJ, McCarroll S, Pato C, Pato MT, Smith AV, NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium, Boehnke M, Scott LJ, Fuchsberger C: Extent to which array genotyping and imputation with large reference panels approximate deep whole-genome sequencing. AMERICAN JOURNAL OF HUMAN GENETICS: 2022; 109:1653-1666.
Grüneis R, Lamina C, Di Maio S, Schönherr S, Zoescher P, Forer L, Streiter G, Peters A, Gieger C, Köttgen A, Kronenberg F, Coassin S: The effect of LPA Thr3888Pro on lipoprotein(a) and coronary artery disease is modified by the LPA KIV-2 variant 4925G>A. ATHEROSCLEROSIS: 2022a; 349:151-159
Schnitzer F, Forer L, Schönherr S, Gieger C, Grallert H, Kronenberg F, Peters A, Lamina C: Association between a polygenic and family risk score on the prevalence and incidence of myocardial infarction in the KORA-F3 study. ATHEROSCLEROSIS: 2022b; 352:10-17
Koschinsky ML, Kronenberg F (editors): Special issue: Lipoprotein(a). ATHEROSCLEROSIS: 2022c; 349: 1-248.
Schäfer T, Kramer K, Werten S, Rupp B, Hoffmeister D: Characterization of the gateway decarboxylase for psilocybin biosynthesis. CHEMBIOCHEM: 2022; 23:e202200551
Kheirkhah A, Lamina C, Kollerits B, Schachtl-Riess JF, Schultheiss UT, Forer L, Sekula P, Kotsis F, Eckardt KU, Kronenberg F, GCKD Investigators: PCSK9 and cardiovascular disease in individuals with moderately decreased kidney function. CLINICAL JOURNAL OF THE AMERICAN SOCIETY OF NEPHROLOGY: 2022; 17:809-818
Kronenberg F, Mora S, Stroes ESG, Ference BA, Arsenault BJ, Berglund L, Dweck MR, Koschinsky M, Lambert G, Mach F, McNeal CJ, Moriarty PM, Natarajan P, Nordestgaard BG, Parhofer KG, Virani SS, von Eckardstein A, Watts GF, Stock JK, Ray KK, Tokgözoğlu LS, Catapano AL: Lipoprotein(a) in atherosclerotic cardiovascular disease and aortic stenosis: a European Atherosclerosis Society consensus statement. EUROPEAN HEART JOURNAL: 2022; 43:3925-3946
Lüth T, Schaake S, Grünewald A, May P, Trinh J, Weissensteiner H: Benchmarking low-frequency variant calling with long-read data on mitochondrial DNA. FRONTIERS IN GENETICS: 2022; 13:887644
Weissensteiner H, Forer L, Fendt L, Kheirkhah A, Salas A, Kronenberg F, Schoenherr S: Contamination detection in sequencing studies using the mitochondrial phylogeny. GENOME RESEARCH: 2021; 31:309-316
Fazzini F, Fendt L, Schönherr S, Forer L, Schöpf B, Streiter G, Losso JL, Kloss-Brandstätter A, Kronenberg F, Weissensteiner H: Analyzing low-level mtDNA heteroplasmy-pitfalls and challenges from bench to benchmarking. INTERNATIONAL JOURNAL OF MOLECULARE SCIENCES: 2021a; 22:935
Cortes-Figueiredo F, Carvalho FS, Fonseca AC, Paul F, Ferro JM, Schönherr S, Weissensteiner H*, Morais VA*: From forensics to clinical research: Expanding the variant calling pipeline for the Precision ID mtDNA Whole Genome Panel. INTERNATIONAL JOURNAL OF MOLECULARE SCIENCES: 2021b; 22:12031, * corresponding authors
Fazzini F, Lamina C, Raftopoulou A, Koller A, Fuchsberger C, Pattaro C, Del Greco FM, Döttelmayer P, Fendt L, Fritz J, Meiselbach H, Schönherr S, Forer L, Weissensteiner H, Pramstaller PP, Eckardt KU, Hicks AA, Kronenberg F: Association of mitochondrial DNA copy number with metabolic syndrome and type 2 diabetes in 14 176 individuals. JOURNAL OF INTERNAL MEDICINE: 2021; 290:190-202
Di Maio S, Lamina C, Coassin S, Forer L, Würzner R, Schönherr S, Kronenberg F: Lipoprotein(a) and SARS-CoV-2 infections: Susceptibility to infections, ischemic heart disease and thromboembolic events. JOURNAL OF INTERNAL MEDICINE: 2022a; 291:101-107
Schwaiger JP, Kollerits B, Steinbrenner I, Weissensteiner H, Schönherr S, Forer L, Kotsis F, Lamina C, Schneider MP, Schultheiss UT, Wanner C, Köttgen A, Eckardt KU, Kronenberg F, GCKD Investigators: Apolipoprotein A-IV concentrations and clinical outcomes in a large chronic kidney disease cohort: Results from the GCKD study. JOURNAL OF INTERNAL MEDICINE: 2022b; 291:622-636
Schachtl-Riess JF, Coassin S, Lamina C, Demetz E, Streiter G, Hilbe R, Kronenberg F: Lysis reagents, cell numbers, and calculation method influence high-throughput measurement of HDL-mediated cholesterol efflux capacity. JOURNAL OF LIPID RESEARCH: 2021; 62:100125
Grüneis R, Weissensteiner H, Lamina C, Schönherr S, Forer L, Di Maio S, Streiter G, Peters A, Gieger C, Kronenberg F, Coassin S: The kringle IV type 2 domain variant 4925G>A causes the elusive association signal of the LPA pentanucleotide repeat. JOURNAL OF LIPID RESEARCH: 2022; 63:100306
Schachtl-Riess JF, Kheirkhah A, Grüneis R, Di Maio S, Schoenherr S, Streiter G, Losso JL, Paulweber B, Eckardt KU, Köttgen A, Lamina C, Kronenberg F, Coassin S: Frequent LPA KIV-2 variants lower lipoprotein(a) concentrations and protect against coronary artery disease. JOURNAL OF THE AMERICAN COLLEGE OF CARDIOLOGY: 2021; 78:437-449
Naschberger A, Baradaran R, Rupp B*, Carroni M*: The structure of neurofibromin isoform 2 reveals different functional states. NATURE: 2021: 599, 315–319, * authors contributed equally
Kloss-Brandstätter A, Summerer M, Horst D, Horst B, Streiter G, Raschenberger J, Kronenberg F, Sanguansermsri T, Horst J, Weissensteiner H: An in-depth analysis of the mitochondrial phylogenetic landscape of Cambodia. SCIENTIFIC REPORTS: 2021; 11:10816
Hämmerle M, Forer L, Schönherr S, Peters A, Grallert H, Kronenberg F, Gieger C, Lamina C: A family and a genome-wide polygenic risk score are independently associated with stroke in a population-based study. STROKE: 2022; 53:2331-2339
Selection of Funding
“Studying an Lp(a) mutation in human RNA and liver organoids”, FWF, Stefan Coassin
“The structural basis of psilocybin biosynthesis” FWF, Bernhard Rupp
“Host response in opportunistic infections”, FWF PhD Programme, Florian Kronenberg
“Imputation Server – BioDATA Catalyst Program”, NIH-III; Schönherr Sebastian
“Lp(a) in der BIS Studie”, with Charité Berlin; Florian Kronenberg
“Apolipoprotein(a) isoforms and apolipoprotein A-IV concentrations in the Rotterdam Study and DiaGene Study”; University of Amsterdam, Florian Kronenberg
“Molecular Investigation of high lipoprotein(a) pedigrees.” Dr. Legerlotz-Stiftung, Silvia Di Maio
“CUC-Striking a new path to understand HDL functionality.” Dr. Legerlotz-Stiftung, Johanna Schachtl-Rieß.
“Askimed-Cloud-Plattform zur Erhebung, Verwaltung und Analyse von Daten im Gesundheitssektor”, Lukas Forer, Sebastian Schönherr
Collaborations
Gonçalo Abecasis, Center of Statistical Genetics, University of Michigan, Ann Arbor, USA
Steven C. Hunt, Cardiovascular Genetics Division, University of Utah, Salt Lake City, USA
Institute of Epidemiology, Helmholtz Zentrum München, Neuherberg, Germany
Kai-Uwe Eckardt, Department of Nephrology and Medical Intensive Care, Charité—Universitätsmedizin Berlin, Berlin, Germany
Nicole Probst-Hensch, Swiss Tropical and Public Health Institute, Basel, Switzerland
Anna Köttgen, Institute of Genetic Epidemiology, University of Freiburg, Freiburg, Germany
Eric Sijbrands, Department of Internal Medicine, Erasmus MC-University Medical Center Rotterdam, The Netherlands
Iris M. Heid, Department of Genetic Epidemiology, University of Regensburg, Regensburg, Germany
Gilles Lambert, Laboratoire INSERM UMR 1188 DéTROI, Université de La Réunion, Sainte Clotilde, France
Devices & Services
Free Next-Generation Genotype Imputation Service using most comprehensive reference genomes: https://imputationserver.sph.umich.edu
mtDNA-Server & new mtDNA analysis tool sets: https://mitoverse.i-med.ac.at
Haplogroup classification service HaploGrep3: https://haplogrep.i-med.ac.at
Next generation eCRF system for clinical studies: https://www.askimed.com/