Innrain 80-82
6020 Innsbruck
Email: Lukas.A.Huber@i-med.ac.at
Website: https://cellbiology.i-med.ac.at/
Research year
Research Branch (ÖSTAT Classification)
106013, 106023, 106037, 106041, 106052, 301904, 302016, 302025, 302035, 302078, 302084
Keywords
cancer, Cell biology, genetic model organisms, metabolic signalling, organelle proteostasis, protein transport and sorting, proteomics, and rare diseases
Research Focus
Rare diseases
Cancer
Membrane Proteostasis and Signalling
Cell Differentiation
Signal Transduction and Proteomics
General Facts
At the Institute of Cell Biology, we are unraveling the molecular mechanisms that govern cellular function, from protein structure to the dysregulation underlying rare diseases. Our independent research groups are addressing fundamental questions in cell biology, structural biology and biochemistry by means of cutting-edge approaches, including high-resolution imaging, cryo-electron microscopy, genome editing and quantitative proteomics. We are using genetic models (yeast, mouse and human cells) to bridge the gap between molecular insights and disease mechanisms and are fostering translational collaborations with clinical and biotech partners. As a hub for innovation, we provide a vibrant, international research environment for Masters students, PhD students and postdoctoral fellows.
We are all funded by the FWF and represent major contributors to the MCBD (Molecular and Cellular Biology of Disease) PhD programme and the DOC82 (Cellular Basis of Diseases) doc.funds initiative, as well as to leading EU-funded projects and global academic partnerships. By bringing together basic research and clinical applications, we are driving discoveries that elucidate cellular health and disease.
Research
The Huber laboratory: unlocking the secrets of lysosomal mTOR signalling
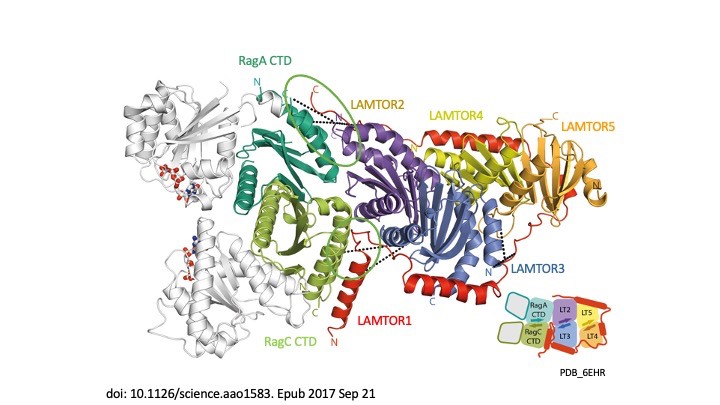
Our story begins in 2003 when we transformed our understanding of cellular signalling by identifying the LAMTOR/Ragulator complex and showing how the p14-MP1 scaffold anchors MAPK signalling to endosomes, a crucial step in regulating cell growth and survival (Teis et al., Dev Cell 2003). In 2007, we linked this complex to human disease, identifying p14 deficiency as the cause of a devastating primary immunodeficiency (Bohn et al, Nat Med 2007). This was the first indication that LAMTOR was more than a mere signalling hub – it is a lifeline. The plot thickened in 2015 when, in collaboration with Giulio Superti-Furga, we discovered SLC38A9, the long-sought amino-acid sensor that allows mTORC1 to ‘taste’ nutrients in lysosomes (Rebsamen et al, Nature 2015). Suddenly the pieces fell into place – LAMTOR was not only a scaffold, it was the control centre of metabolic decision-making. The structural revolution enabled us in 2017 to capture the first crystal structure of LAMTOR/Ragulator with Rag GTPases, revealing how this molecular machine aligns mTORC1 signalling on lysosomes (Araujo et al., Science 2017). In the same year, we showed that LAMTOR also controls lysosomal positioning, acting as a brake on endosomal motility (Filipek et al., JCB 2017). The story took an exciting turn in 2020, when – together with Jim Hurley and Andrea Ballabio – we linked LAMTOR dysfunction to the Birt-Hogg-Dubé syndrome, showing how faulty mTORC1 regulation drives this rare disease (Napolitano et al., Nature 2020). Finally, in 2023, we unveiled the megacomplex – a gigantic assembly of mTORC1, TFEB, Ragulator and Rags – painting a complete picture of how lysosomes orchestrate cellular metabolism (Cui et al., Nature 2023). From cancer to immune disorders, the LAMTOR complex is at the heart of disease and we are gradually unlocking its secrets, one discovery at a time.
The Vogel laboratory: intracellular cargo transport in congenital intestinal diseases
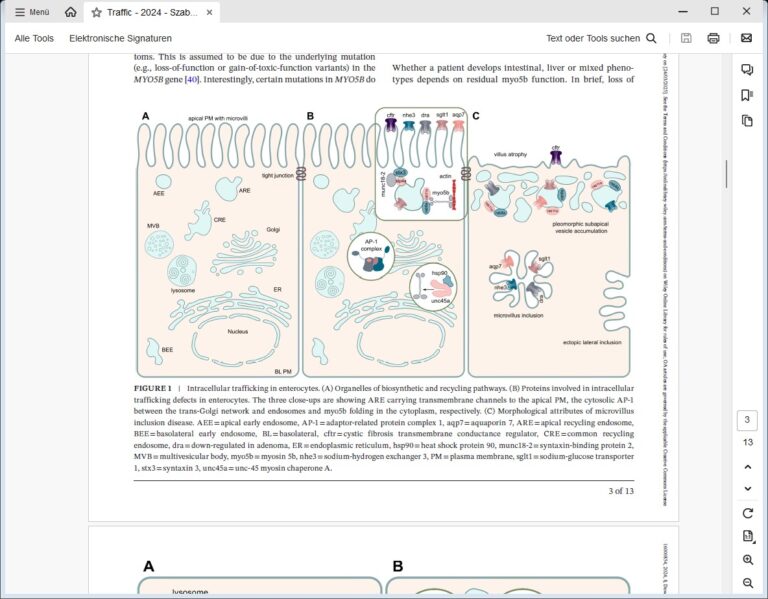
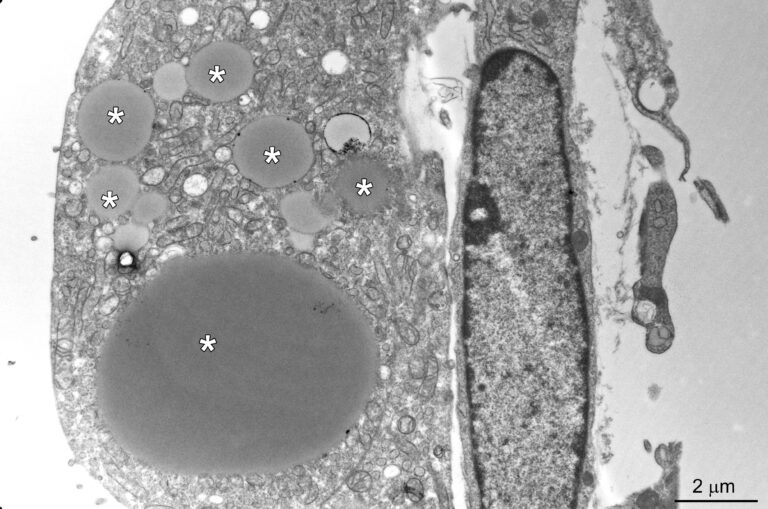
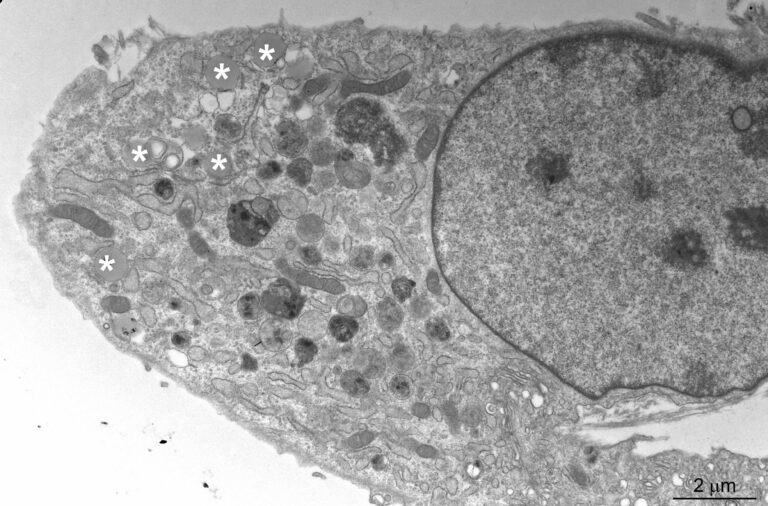
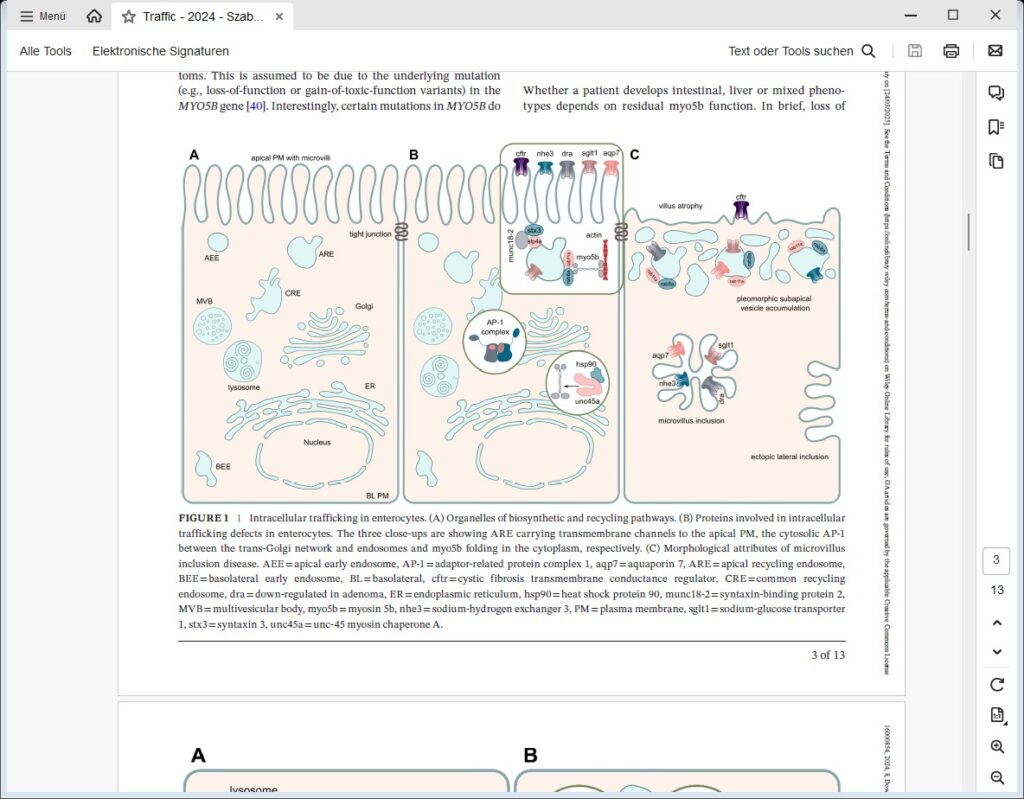
We are focusing on congenital intestinal and liver diseases, which are often caused by mutations in genes that are essential for correct intestinal-epithelial polarization. For example, we have identified mutations in MYO5B and STX3 in microvillus inclusion disease: both genes are essential for the polarized apical cargo transport of transmembrane transporters (Vogel GF et al., JCB, 2015; Vogel GF et al., JCI Insight, 2017; Szabó L et al., Traffic, 2024). We are now turning our attention to myo5b-associated cholestatic liver disease and myo5b-independent apical transport in intestinal epithelial cells.
We recently described the first case of a congenital enteropathy due to mutations in ezrin (manuscript in press), a protein involved in the establishment of proper apical polarity and plasma membrane composition in epithelial cells. We are also addressing the role of the retriever complex and the poorly characterized protein TM9SF4 in polarized plasma membrane cargo trafficking in epithelial cells.
The Vietor laboratory: regulatory mechanisms of cell differentiation
Our group is interested in the roles of the TPA-induced sequence-7 (TIS7, also known as IFRD1) protein and its orthologue SKMc15 (IFRD2). We have shown that they are involved in the regulation of the differentiation of neurons, enterocytes, myocytes and adipocytes and that TIS7 is a transcriptional co-regulator (Lammirato et al., BMC Biol, 2016). We have generated TIS7 and SKMc15 double knockout (dKO) mice and used them to identify a regulatory role for TIS7 in adipocyte differentiation and in resistance to high-fat diet (Vietor et al. eLife, 2023). As dKO mice are substantially leaner, we propose that TIS7 and SKMc15 are involved in physiological adipocyte differentiation. However, our mouse model does not enable us to distinguish between a systemic and a tissue-specific role of the two genes. We are thus generating constitutive knockout mouse models that specifically lack the expression of these genes in either the small intestine or adipocyte tissues. We are also establishing mouse organoid technology to limit the use of experimental animals and to permit us to study the role of the two genes more closely.
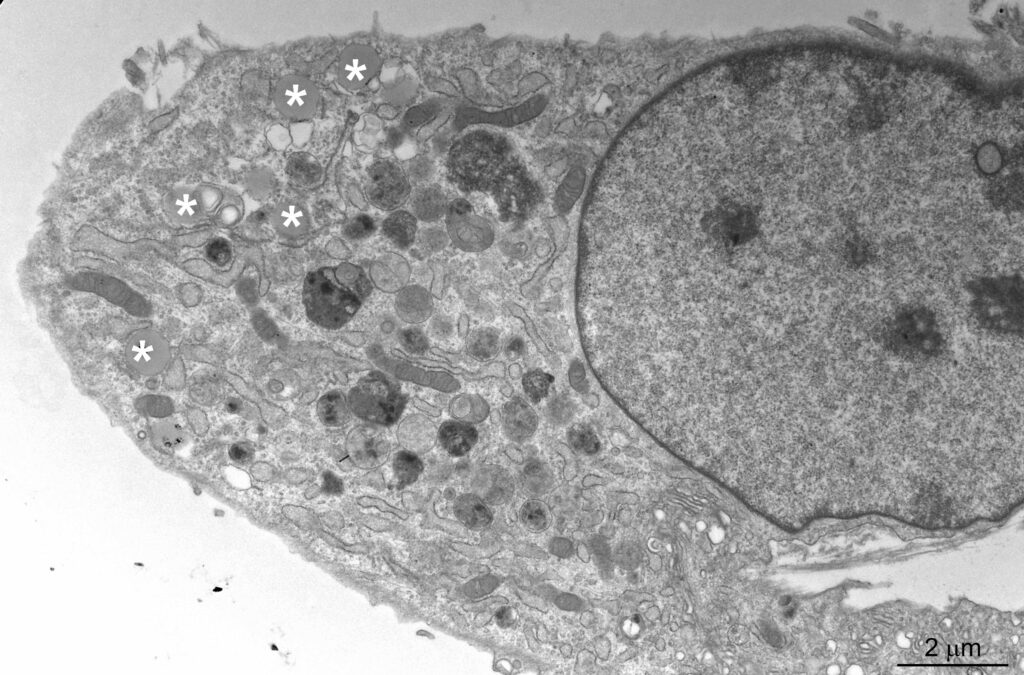
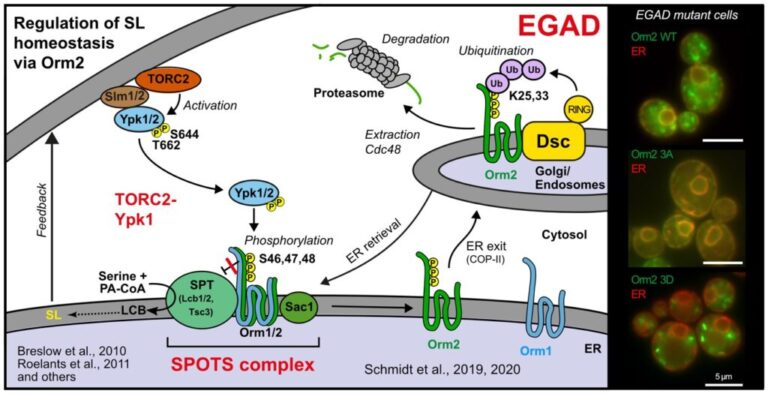
The Schmidt laboratory: regulation of cellular lipid homeostasis
Sphingolipids are an essential class of lipids. Their biosynthesis is initiated at the endoplasmic reticulum by the enzyme serine-palmitoyl-coenzyme A transferase (SPT), the activity of which is controlled by the evolutionarily conserved ORMDL protein family. ORMDL proteins associate with and thereby inhibit SPT. Genetic mutations that affect the interaction are of high pathological relevance and uncontrolled synthesis of sphingolipid precursors causes severe neurodegenerative phenotypes, including amyotrophic lateral sclerosis and frontotemporal dementia. We are studying the regulation of SPT in the light of differences between ORMDL paralogs.
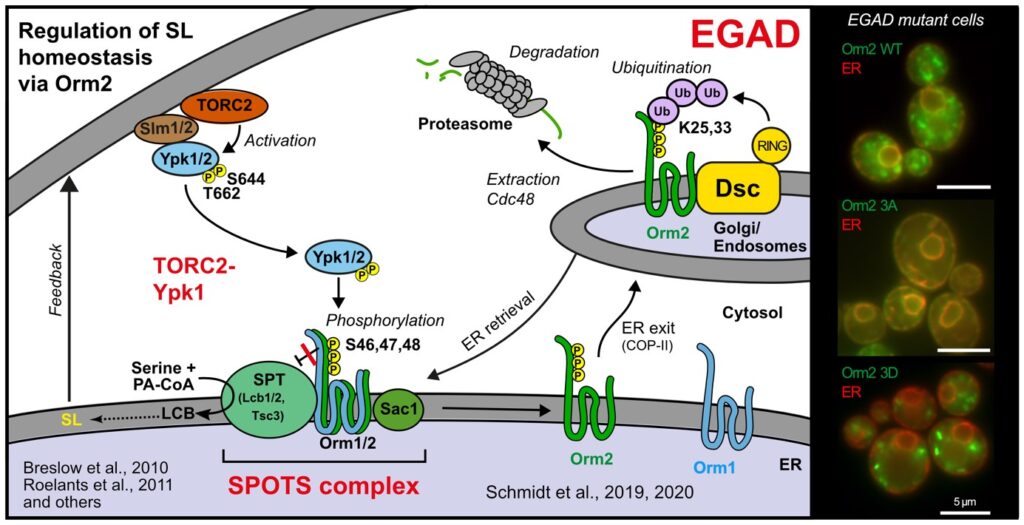
The Valovka laboratory: molecular basis of rare genetic disorders
Rare diseases are often the result of harmful genetic variants but the precise molecular effects of these variants remain poorly understood. Our group is working to identify the genetic culprits behind the disorders and to uncover how they hijack cellular processes to cause disease. In close collaboration with the Department of Paediatrics and Adolescent Medicine at MUI (Andreas Janecke and Thomas Müller), we have identified and are studying unique pathogenic mutations in various genes, including PCNT, SLC5A1, PCSK1, RIPK1 and UNC45A.
The osteo-oto-hepato-enteric (O2HE) syndrome is an autosomal recessive disease ascribed to loss-of-function mutations in the Unc-45 myosin chaperone A (UNC45A) gene. The clinical spectrum includes bone fragility, hearing loss, cholestasis, neurological symptoms and life-threatening diarrhoea associated with microvillus inclusion disease (MVID)-like enteropathy. We are trying to understand how a deficiency of or specific mutations in UNC45A affect the complex molecular mechanism of this severe neonatal disease. We have described a novel missense variant of UNC45A, c.710T>C, with unique pathogenicity caused by altered
chaperone-myosin interactions rather than a complete loss of chaperone function. The findings indicate the possibility of treatment and may guide the development of targeted therapies.
We are also screening natural anti-inflammatory compounds. In collaboration with the Austrian Drug Screening Institute (ADSI), we have developed a screening platform to identify and evaluate bioactive compounds in natural extracts. We were able to show the anti-inflammatory and cytoprotective polypharmacology of the complex herbal medicine Canephron N® (Bionorica SE) and to identify its mechanism of action.
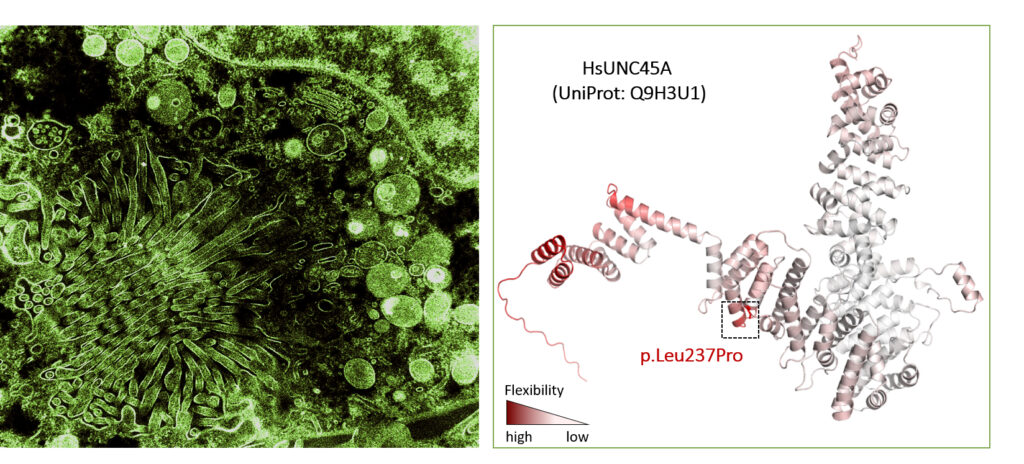
The Araujo laboratory: lysosomal signaling complexes – coordinating organelle function with cellular homeostasis
Lysosomes are membrane-bound organelles with several functions, from protein degradation to the control of cellular metabolism. Due to their importance for proteostasis and signaling, cells have developed mechanisms to control lysosomal numbers, size, movement, degradative capacity, interaction with other organelles and even lysophagy. Dysfunction is linked to numerous diseases, including lysosomal storage disorders, neurodegeneration, metabolic conditions and cancer.
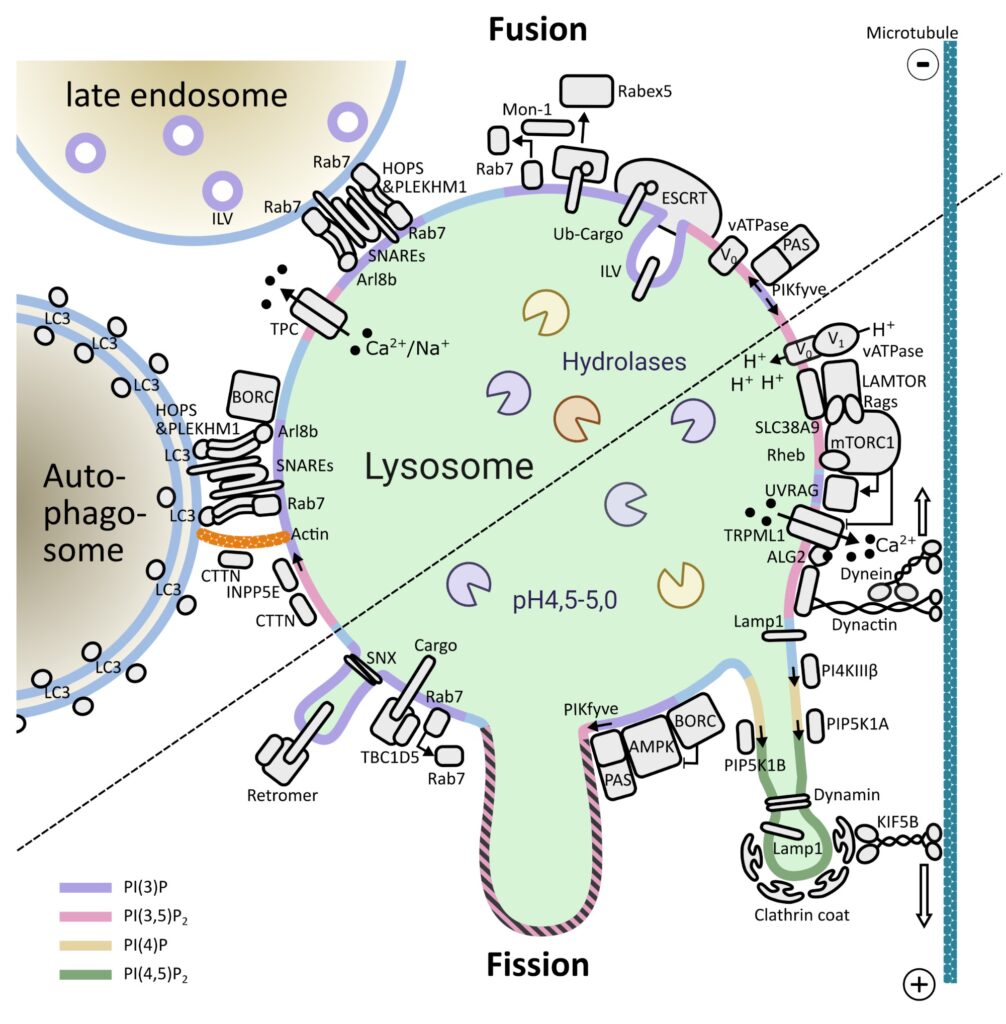
One of the key signaling complexes in the lysosomal membrane is mTORC1. Building on the knowledge from numerous laboratories over more than 30 years of research on this kinase, we are investigating how mTORC1 signaling components bind and coordinate their functions with other lysosomal proteins/complexes to maintain cellular homeostasis. We are also exploring how dysfunction of these processes contributes to disease. We use a combination of classical biochemical and cell biology methods, genome editing, live-cell imaging and super-resolution microscopy.
The Stasyk laboratory: molecular architecture of native lysosomal protein assemblies
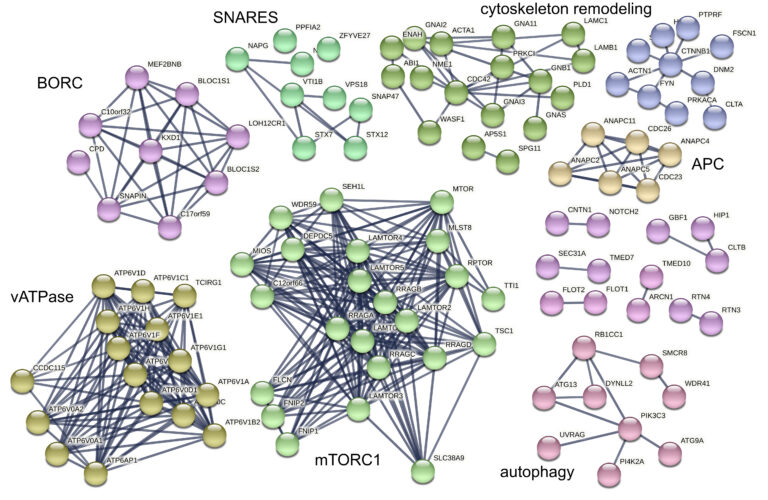
Lysosomes are intracellular signaling platforms that govern cell growth, division and differentiation. Although our knowledge of the molecular mechanisms of the sensing of essential nutrients has advanced significantly, we still lack information on the interplay between the numerous components of sensing and signaling machineries. The LAMTOR/Ragulator complex is a crucial regulator of lysosomal mTORC1, MAPK and AMPK signaling. Via the BORC complex, LAMTOR also coordinates lysosome biogenesis. The existence of distinct LAMTOR complexes on lysosomes raises questions on how anabolic and catabolic signaling, lysosomal biogenesis and positioning are coordinated.
We are aiming to identify all LAMTOR- and BORC-associated protein assemblies on intact lysosomes and to obtain evidence for direct and endogenous protein-protein interactions. We are applying cross-linking mass spectrometry (XL-MS) in combination with subcellular fractionation to understand the molecular architectures of the protein complexes under different physiological conditions. Our approach captures native endogenous protein interactions using a chemical cross-linker that is reactive towards specific amino-acid side chains that are in close spatial proximity.
Pictures
Selected Publications
The Huber Laboratory: Lysosomal mTOR Signalling – The Crossroads between Signal Transduction and Endosomal Biogenesis
1: Vogel GF, Klee KMC, Demir AM, Garczarczyk-Asim D, Hess MW, Huber LA, Mülller T, Janecke AR. Congenital enteropathy caused by ezrin deficiency. Hum Genet.2025 Mar 26. doi: 10.1007/s00439-025-02738-w. Epub ahead of print. PMID:40137958.
2: Waich S, Kreidl K, Vodopiutz J, Demir AM, Pollio AR, Dostál V, Pfaller K, Parlato M, Cerf-Bensussan N, Adam R, Vogel GF, Uhlig HH, Ruemmele FM, Müller T, Hess MW, Janecke AR, Huber LA, Valovka T. Altered chaperone-nonmuscle myosin II interactions drive pathogenicity of the UNC45A c.710T>C variant in osteo-oto-hepato-enteric syndrome. JCI Insight. 2025 Mar 24;10(6):e185508. doi:
10.1172/jci.insight.185508. PMID: 40125554.
3: Liebscher G, Vujic N, Schreiber R, Heine M, Krebiehl C, Duta-Mare M, Lamberti G, de Smet CH, Hess MW, Eichmann TO, Hölzl S, Scheja L, Heeren J, Kratky D, Huber LA. The lysosomal LAMTOR / Ragulator complex is essential for nutrient homeostasis in brown adipose tissue. Mol Metab. 2023 May;71:101705. doi: 10.1016/j.molmet.2023.101705. Epub 2023 Mar 11. PMID: 36907508; PMCID:
PMC10074977.
4: Cui Z, Napolitano G, de Araujo MEG, Esposito A, Monfregola J, Huber LA, Ballabio A, Hurley JH. Structure of the lysosomal mTORC1-TFEB-Rag-Ragulator megacomplex. Nature. 2023 Feb;614(7948):572-579. doi: 10.1038/s41586-022-05652-7. Epub 2023 Jan 25. PMID: 36697823; PMCID: PMC9931586.
5: Vietor I, Cikes D, Piironen K, Vasakou T, Heimdörfer D, Gstir R, Erlacher MD,Tancevski I, Eller P, Demetz E, Hess MW, Kuhn V, Degenhart G, Rozman J,Klingenspor M, Hrabe de Angelis M, Valovka T, Huber LA. The negative adipogenesis regulator is transcriptionally regulated by
(TIS7) and translationally by its orthologue (SKMc15).
Elife. 2023 Aug 21;12:e88350. doi: 10.7554/eLife.88350. PMID: 37603466; PMCID: PMC10468205.
The Vietor Laboratory: Regulatory Mechanisms of Cell Differentiation
An Overview of Autophagy in Hematopoietic Stem Cell Transplantation.
Montazersaheb S, Ehsani A, Fathi E, Farahzadi R, Vietor I. Front Bioeng Biotechnol. 2022 May 23;10:849768. doi: 10.3389/fbioe.2022.849768. eCollection 2022. PMID: 35677295.
Vietor I, Cikes D, Piironen K, Vasakou T, Heimdörfer D, Gstir R, Erlacher MD, Tancevski I, Eller P, Demetz E, Hess MW, Kuhn V, Degenhart G, Rozman J, Klingenspor M, Hrabe de Angelis M, Valovka T, Huber LA.Elife. 2023 Aug 21;12:e88350. doi: 10.7554/eLife.88350.PMID: 37603466.
Farahzadi R, Fathi E, Mesbah-Namin SA, Vietor I. Regen Ther. 2023 May 3;23:94-101. doi: 10.1016/j.reth.2023.04.004. eCollection 2023 Jun. PMID: 37206538.
Fathi E, Montazersaheb S, Vandghanooni S, Farahzadi R, Vietor I.Curr Mol Med. 2023;23(3):266-274. doi: 10.2174/1566524022666220118103136.PMID: 35040412
Fathi E, Mesbah-Namin SA, Vietor I, Farahzadi R.Iran J Basic Med Sci. 2022 Oct;25(10):1222-1227. doi: 10.22038/IJBMS.2022.66737.14633.PMID: 36311196
Fathi E, Montazersaheb S, Sanaat Z, Nakhlband A, Vandghanooni S, Farahzadi R, Vietor I.Curr Stem Cell Res Ther. 2023;18(2):231-236. doi: 10.2174/1574888X17666220511141123.PMID: 35546751
Fathi E, Azarbad S, Farahzadi R, Javanmardi S, Vietor I.Curr Gene Ther. 2022;22(2):152-161. doi: 10.2174/1566523221666210519111933.PMID: 34011256
The Vogel Laboratory: Intracellular Cargo Transport in Congenital Intestinal Diseases
Schlieben LD, Achleitner MT, Bourke B, Diesner M, Feichtinger RG, Fichtner A, Flechtenmacher C, Hadzic N, Hegarty R, Heilos A, Janecke A, Konstantopoulou V, Lenz D, Mayr JA, Müller T, Prokisch H, Vogel GF. Hepatol Commun. 2024 Nov 29;8(12):e0598. doi: 10.1097/HC9.0000000000000598. eCollection 2024 Dec 1.
Intracellular Trafficking Defects in Congenital Intestinal and Hepatic Diseases.
Szabó L, Pollio AR, Vogel GF. Traffic. 2024 Aug;25(8):e12954. doi: 10.1111/tra.12954.
Vogel GF, Podpeskar A, Rieder D, Salzer H, Garczarczyk-Asim D, Wang L, Abuduxikuer K, Wang JS, Scharrer A, Faqeih EA, Aseeri AT, Vodopiutz J, Heilos A, Pichler J, Huber WD, Müller T, Knisely AS, Janecke AR. Clin Genet. 2024 Sep;106(3):224-233. doi: 10.1111/cge.14524. Epub 2024 Mar 29.
Natural history of Wolcott-Rallison syndrome: A systematic review and follow-up study.
Aldrian D, Bochdansky C, Kavallar AM, Mayerhofer C, Deeb A, Habeb A, Romera Rabasa A, Khadilkar A, Uçar A, Knoppke B, Zafeiriou D, Lang-Muritano M, Miqdady M, Judmaier S, McLin V, Furdela V, Müller T, Vogel GF.
Efficacy of Elimination Diets in Eosinophilic Esophagitis: A Systematic Review and Meta-analysis.
Mayerhofer C, Kavallar AM, Aldrian D, Lindner AK, Müller T, Vogel GF.Clin Gastroenterol Hepatol. 2023 Aug;21(9):2197-2210.e3. doi: 10.1016/j.cgh.2023.01.019. Epub 2023 Jan 31.PMID: 36731591.
Klee KMC, Hess MW, Lohmüller M, Herzog S, Pfaller K, Müller T, Vogel GF, Huber LA. Elife. 2023 Jan 20;12:e80135. doi: 10.7554/eLife.80135. PMID: 36661306.
The Schmidt Laboratory: Regulation of cellular lipid homeostasis
The structure of the Orm2-containing serine palmitoyltransferase complex reveals distinct inhibitory potentials of yeast Orm proteins. Körner C, Schäfer JH, Esch BM, Parey K, Walter S, Teis D, Januliene D, Schmidt O*, Moeller A*, Fröhlich F*. Cell Rep. 2024 Aug 20;43(8):114627. doi: 10.1016/j.celrep.2024.114627. PMID: 39167489 (* shared corresponding authors)
Weyer Y, Schwabl SI, Tang X, Purwar A, Siegmann K, Ruepp A, Dunzendorfer-Matt T, Widerin MA, Niedrist V, Mutsters NJM, Tettamanti MG, Weys S, Sarg B, Kremser L, Liedl KR, Schmidt O, Teis D. Nat Commun. 2024 Oct 26;15(1):9257. doi: 10.1038/s41467-024-53676-6. PMID: 39461958
A metabolically controlled contact site between vacuoles and lipid droplets in yeast.
Diep DTV, Collado J, Hugenroth M, Fausten RM, Percifull L, Wälte M, Schuberth C, Schmidt O, Fernández-Busnadiego R, Bohnert M.Dev Cell. 2024 Mar 25;59(6):740-758.e10. doi: 10.1016/j.devcel.2024.01.016. Epub 2024 Feb 16.PMID: 38367622
Targeting the CLK2/SRSF9 splicing axis in prostate cancer leads to decreased ARV7 expression.
Van Goubergen J, Peřina M, Handle F, Morales E, Kremer A, Schmidt O, Kristiansen G, Cronauer MV, Santer FR.Mol Oncol. 2025 Feb;19(2):496-518. doi: 10.1002/1878-0261.13728. Epub 2024 Sep 11.PMID: 39258426
Identification of distinct active pools of yeast serine palmitoyltransferase in sub-compartments of the ER.
Esch BM, Walter S, Schmidt O, Fröhlich F.J Cell Sci. 2023 Dec 1;136(23):jcs261353. doi: 10.1242/jcs.261353. Epub 2023 Dec 19.PMID: 37982431
The Valovka Laboratory: Molecular Basis of Rare Genetic Disorders
Vietor I, Cikes D, Piironen K, Vasakou T, Heimdörfer D, Gstir R, Erlacher MD, Tancevski I, Eller P, Demetz E, Hess MW, Kuhn V, Degenhart G, Rozman J, Klingenspor M, Hrabe de Angelis M, Valovka T, Huber LA. Elife. 2023 Aug 21;12:e88350. doi: 10.7554/eLife.88350. PMID: 37603466
SLC5A1 Variants in Turkish Patients with Congenital Glucose-Galactose Malabsorption.
Hoşnut FÖ, Janecke AR, Şahin G, Vogel GF, Lafcı NG, Bichler P, Müller T, Huber LA, Valovka T, Aksu AÜ. Genes (Basel). 2023 Jun 27;14(7):1359. doi: 10.3390/genes14071359. PMID: 37510265
Pathogenic Deep Intronic PCSK1 Variant Causes Proprotein Convertase 1/3 Deficiency in a Family.
Huber LM, Subaşıoğlu A, Garczarczyk-Asim D, Valovka T, Müller T, Adam R, Janecke AR.Clin Genet. 2025 Feb 1. doi: 10.1111/cge.14717. Online ahead of print.PMID: 39891480
Milosevic M, Magnutzki A, Braun T, Hussain S, Jakschitz T, Kragl M, Soeberdt M, Nausch B, Bonn GK, Huber LA, Valovka T.Biomed Pharmacother. 2025 Jan;182:117747. doi: 10.1016/j.biopha.2024.117747. Epub 2024 Dec 12.PMID: 39671726
Waich S, Kreidl K, Vodopiutz J, Demir AM, Pollio AR, Dostál V, Pfaller K, Parlato M, Cerf-Bensussan N, Adam R, Vogel GF, Uhlig HH, Ruemmele FM, Müller T, Hess MW, Janecke AR, Huber LA, Valovka T. JCI Insight. 2025 Mar 24;10(6):e185508. doi: 10.1172/jci.insight.185508. PMID: 40125554
The Araujo Laboratory: Lysosomal signaling complexes – Coordinating organelle function with cellular homeostasis
Structure of the lysosomal mTORC1-TFEB-Rag-Ragulator megacomplex.
Cui Z, Napolitano G, de Araujo MEG, Esposito A, Monfregola J, Huber LA, Ballabio A, Hurley JH.Nature. 2023 Feb;614(7948):572-579. doi: 10.1038/s41586-022-05652-7. Epub 2023 Jan 25.PMID: 36697823
Sambri I, Ferniani M, Campostrini G, Testa M, Meraviglia V, de Araujo MEG, Dokládal L, Vilardo C, Monfregola J, Zampelli N, Vecchio Blanco FD, Torella A, Ruosi C, Fecarotta S, Parenti G, Staiano L, Bellin M, Huber LA, De Virgilio C, Trepiccione F, Nigro V, Ballabio A.Nat Commun. 2023 May 15;14(1):2775. doi: 10.1038/s41467-023-38428-2.PMID: 37188688
Heimdörfer D, Vorleuter A, Eschlböck A, Spathopoulou A, Suarez-Cubero M, Farhan H, Reiterer V, Spanjaard M, Schaaf CP, Huber LA, Kremser L, Sarg B, Edenhofer F, Geley S, de Araujo MEG, Huettenhofer A. Am J Hum Genet. 2024 Jul 11;111(7):1383-1404. doi: 10.1016/j.ajhg.2024.05.023. Epub 2024 Jun 21. PMID: 38908375
The Stasyk Laboratory: Molecular architecture of native lysosomal protein assemblies
DIGE-Based Phosphoproteomic Analysis.
Stasyk T, Huber LA.Methods Mol Biol. 2023;2596:97-104. doi: 10.1007/978-1-0716-2831-7_7.PMID: 36378433
Selection of Funding
The Huber Laboratory: Lysosomal mTOR Signalling – The Crossroads Between Signal Transduction and Endosomal Biogenesis
FWF P 32608 and P 26682, 10.55776/PAT2389124 and FWF DOC 82 doc.fund Cellular Basis of Diseases: Molecular Control of Metabolism and Inflammation
Organelle proteostasis in cellular quiescence and growth SUB4 FWF Grant DOI:10.55776/FG20
New functions of the KICSTOR component SZT2 FWF Grant DOI: 10.55776/PAT2389124
The Vogel Laboratory: Intracellular Cargo Transport in Congenital Intestinal Diseases
ÖNB Anniversary Fund 18019, TWF Grant 0404/2386, ÖGKJ research grant
The Vietor Laboratory: Regulatory Mechanisms of Cell Differentiation
IMPULSE Iran Austria
FWF P36861-B
The Schmidt laboratory: Regulation of cellular lipid homeostasis
Lipotype – Lipid excellence award 2019 (#2)
FWF P36187-B “individual functions of Orm/ORMDL family proteins in membrane homeostasis”
The Stasyk Laboratory: Molecular architecture of native lysosomal protein assemblies
FWF P36975 – Molecular architecture of native LAMTOR and BORC assemblies
The Araujo Laboratory: Lysosomal signalling complexes – Coordinating organelle function with cellular homeostasis
Organelle proteostasis in cellular quiescence and growth SUB5
FWF Grant DOI:10.55776/FG20
TWF GZ F16676/5-2019 – A potential new mechanism for selective modulation of mTORC1
Collaborations
The Huber Laboratory: Lysosomal mTOR Signalling – The Crossroads between Signal Transduction and Endosomal Biogenesis
Giulio Suerti-Furga CeMM, David Haselbach IMP, Manuela Baccarini Max Perutz Laboratories all Vienna Austria
Dagmar Kratky Gottfried Schatz Research Center, Molecular Biology and Biochemistry, Medical University of Graz, AUT
Andrea Ballabio TIGEM, Naples, Italy
Structural Membrane Biology, Jinm Hurley University of California, Berkely, US
The Vogel Laboratory: Intracellular Cargo Transport in Congenital Intestinal Diseases
Saskia Wortmann, Paediatrics, University Hospital Salzburg, AT
The Vietor Laboratory: Regulatory Mechanisms of Cell Differentiation
Ezzatollah Fathi, Department of Clinical Sciences, Faculty of Veterinary Medicine, University of Tabriz, Tabriz, Iran
The Schmidt laboratory: Regulation of cellular lipid homeostasis
Florian Fröhlich, Osnabrück University, DE
Maria Bohnert, Münster University, DE
Robbie Loewith, University of Geneva, CH
Matthias Peter, ETH Zurich, CH
The Valovka Lab: Molecular Basis of Rare Genetic Disorders
Julia Vodopiutz, Department of Pediatrics and Adolescent Medicine, Medical University of Vienna, AUT
Daniel Kotlarz, Department of Pediatrics, Ludwig-Maximilians-Universität (LMU) Munich, DE Christoph Griesbeck, Department of Biotechnology and Food Engineering, MCI – The Entrepreneurial School, AUT
ADSI-Austrian Drug Screening Institute, AUT
Bionorica SE, DE
The Araujo Laboratory: Lysosomal signaling complexes – Coordinating organelle function with cellular homeostasis
Gennaro Napolitano (Tigem, Naples, IT)
David Haselbach (IMP, Vienna, AUT)
Kathrin Thedieck (Institute of Biochemistry,University of Innsbruck, AUT)
The Stasyk Laboratory: Molecular architecture of native lysosomal protein assemblies
Luca Fava (Armenise-Harvard Laboratory of Cell Division, CIBIO Department, University of Trento, IT)