Schöpfstraße 3
6020 Innsbruck
Email: Florian.Kronenberg@i-med.ac.at
Website: https://genepi.i-med.ac.at/
Research year
Research Branch (ÖSTAT Classification)
102004, 102010, 106014, 301301, 301302, 303007
Keywords
Research Focus
We aim to identify the determinants of health and disease related to genetic variability, environmental factors and biochemical parameters and study their physiological or pathophysiological functions.
The phenotypes of interest to us are complex due to the interplay of these factors and are related to conditions such as atherosclerosis, diabetes mellitus, metabolic syndrome, chronic kidney disease and cancer, as well as associated intermediate phenotypes such as lipoprotein metabolism and inflammation. We are particularly interested in genetic and non-genetic factors that influence lipoprotein(a) concentrations.
Our institute serves as a bridge between basic and clinical research. We work with genetic epidemiological methods, including association studies of candidate genes, as well as hypothesis-free genome-wide association studies and clinical epidemiological methods. For functional studies, we use a variety of cell culture models and implement state-of-the-art methods in protein chemistry and molecular biology. Finally, our institute is highly interested and experienced in developing computational approaches for medical genomic research and diagnosis.
General Facts
The institute relocated to Schöpfstraße 3 in early 2025. Consisting of 20–25 members, it serves as a bridge between basic and clinical research.
Our strength lies in our interconnected collaboration across three areas: 1) A protein chemistry and cell culture laboratory conducts various structural, functional and epidemiological studies on different phenotypes related to lipoprotein metabolism and other metabolic traits (e.g. Lp(a), apolipoprotein A-IV, afamin and PCSK9), as well as cholesterol efflux assays; 2) A molecular genetics laboratory performs sequencing and genotyping for various projects, focusing on mitochondrial DNA and complex genetic regions such as the K-IV type 2 region of the LPA gene. New technologies, such as nanopore sequencing, have recently become a major focus of this research; 3) The computational genomics and statistical genetics unit focuses on statistics, epidemiology, computer science and bioinformatics. This unit serves as an important link between the various research groups. The close interconnection of these three units, forming a strongly collaborative alliance has become increasingly evident in recent years. There are barely any major projects involving fewer than two of these main groups. The institute’s output and success are based on constant dialogue between the various disciplines and a critical, problem-oriented elucidation of research questions.
Research
Lipoprotein(a)
Stefan Coassin, Sebastian Schönherr, Silvia Di Maio, Claudia Lamina, Florian Kronenberg
High concentrations of lipoprotein(a) (Lp(a)) are a significant risk factor for cardiovascular disease. Lp(a) levels are regulated almost entirely by the intricate interplay of hundreds of genetic variants in the LPA gene. Our research aims to unravel the complex genetic regulation of Lp(a) and investigate its association with disease.
- Nanopore sequencing in complex genome regions
We use nanopore sequencing and other single-molecule technologies to analyse difficult genomic regions.

The LPA gene, and specifically the KIV-2 VNTR (Variable Number Tandem Repeats) within it, is one of the most complex regions of the genome. Recently, we developed a nanopore sequencing approach incorporating unique molecular identifiers, which reduces sequencing error rates by three orders of magnitude and enables virtually error-free single-molecule sequencing (Genome Med. 2024). This approach provides unprecedented insights into KIV-2 mutation patterns and evolution. This technology has a wide range of applications and can be used to answer any research question that requires error free, quantitative, single-molecule sequences. Examples include viral sequencing, metagenomics and the detection of low-level oncogenic mutations. This work is funded by the new FWF project 10.55776/PAT5152823 (2024-2028), led by Stefan Coassin.
- Uncovering Missing Heritability in Large-Scale Biobank Data
We are developing innovative methods to detect genetic variations in complex regions of the human genome known as VNTRs. These regions are frequently overlooked by standard sequencing techniques and thus largely missed in GWAS. Using the LPA gene as a model, we employ advanced computational approaches to accurately identify variations within these repetitive DNA sequences.

We have already applied our method to data from the 1000 Genomes Project and the UK Biobank, which includes around 500,000 samples. Our vision is to apply this approach to other similar genomic regions, enabling the discovery of significant medical information previously hidden (Genome Biol. 2024).
- Lp(a) as a risk factor for cardiovascular diseases in high-risk patient groups
Although Lp(a) is a recognised risk factor for cardiovascular disease, there is limited evidence of this relationship in patients with chronic kidney disease (CKD), which is a major risk factor in its own right. In a prospective cohort study with more than 5,000 CKD patients, we demonstrated that elevated Lp(a) concentrations and genetic determinants of Lp(a), reflecting lifelong unconfounded effects, were significantly associated with cardiovascular disease at baseline and during follow-up. Our findings imply that Lp(a) should be considered an additional risk factor for this group of patients (J. Internal Med 2024).
Follow-up initiatives on the Lp(a) Consensus paper of the European Atherosclerosis Society (EAS)
Florian Kronenberg
The publication of the Lp(a) consensus paper in 2022, as well as the development of Lp(a)-lowering drugs in phase 3 clinical trials, created enormous momentum, resulting in numerous publications in this field. As a consensus group, we have decided to publish a further paper containing answers to 30 frequently asked questions on practical issues related to epidemiological aspects, risk assessment, measurement issues and the management of high Lp(a) concentrations (Atherosclerosis 2023a). Furthermore, three reviews on this topic have been published (Pharmacol. Res. 2023; Curr. Atherosclerosis Rep. 2024; JAMA 2025)
The Lp(a) International Task Force was established under the umbrella of the FH Europe Foundation and is chaired by Florian Kronenberg. The task force has developed an ambitious five-year strategy and roadmap to make Lp(a) measurement the global norm and ensure that elevated Lp(a) is managed equitably to help prevent cardiovascular disease and related deaths. The first Global Summit on Lp(a), held in March 2025 to coincide with Lp(a) Awareness Day brought together key opinion leaders, policymakers and individuals with personal experience of Lp(a) from all over the world. The first cost-effectiveness analysis of Lp(a) measurement was presented during this summit and the “Brussels International Declaration on Lp(a) Testing and Management” will influence policy discussions on the importance of measuring elevated Lp(a) at European and international levels. This declaration has been recently published (Atherosclerosis 2025) can be endorsed here: https://fhef.org/brussels-international-declaration/
Raising awareness of Lp(a) became an important aspect, resulting in more than 100 presentations during the reporting period, mostly at international meetings around the globe as well as webinars with a wide outreach. Furthermore, we launched a website to inform healthcare providers as well as the general public on Lp(a) (https://genepi.i-med.ac.at/lpa/).
Biomarkers
Claudia Lamina, Barbara Kollerits, Johanna Schachtl-Rieß, Florian Kronenberg
Apolipoprotein A-IV (apoA-IV) is a 46 kDa glycoprotein involved in reverse cholesterol transport. It exhibits antiatherogenic, antioxidative, antithrombotic and antiinflammatory properties, with levels increasing significantly during the kidney disease (CKD). Afamin is a vitamin E-binding glycoprotein that is primarily expressed in the liver and kidneys. In two large, pooled analyses of up to 20,000 participants, we demonstrated that afamin plasma concentrations predict the prevalence and incidence of metabolic syndrome and diabetes mellitus.
- Apolipoprotein A-IV and cancer
CKD is strongly associated with inflammation and oxidative stress, which favour the development of cancer. Proteomics studies have identified a link between low apoA-IV and various forms of cancer. Data from the GCKD study, which involved a large cohort of CKD patients, indicate that high concentrations of apoA-IV are associated with a lower incidence of fatal and non-fatal cancer events (BMC Cancer 2024).
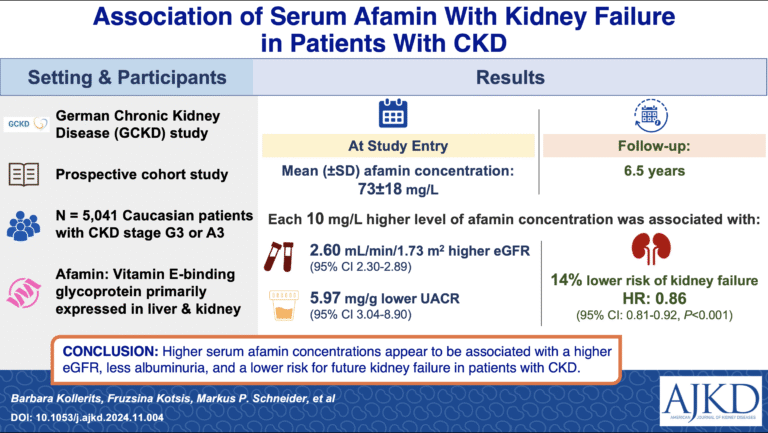
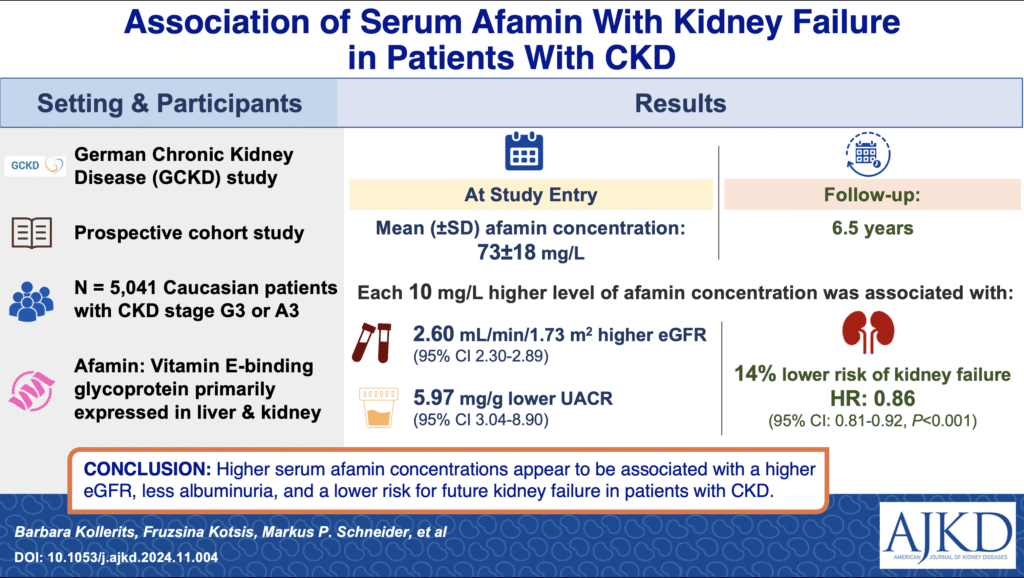
- Afamin and its association with kidney function
In the first prospective study of over 5,000 patients with Chronic Kidney Disease (CKD) (GCKD study), we found a highly significant correlation between serum afamin concentrations and parameters of kidney function, as well as a lower risk of future kidney failure. Thus, afamin may indicate aspects of kidney function beyond changes to the filtration barrier (Am J. Kidney Dis. 2025).
- GWAS on PCSK9 concentrations
The largest meta-analysis of Genome-Wide Association Studies (GWAS) was performed on PCSK9 concentrations in white populations (n=20,579). Five novel loci were identified that were genome-wide significantly associated with PCSK9 concentrations. We also confirmed the loci that were previously identified as being genome-wide significant (Atherosclerosis, 2023b).
- GWAS on HDL-mediated cholesterol efflux capacity
Epidemiological studies have shown that the capacity of macrophages to efflux cholesterol (CEC) mediated by HDL, is inversely associated with cardiovascular disease and may protect against atherosclerosis. In order to identify the genetic and non-genetic determinants of CEC, we measured it in the German Chronic Kidney Disease (GCKD) study (n=5,171). HDL cholesterol and triglyceride concentrations were the most important variables associated with CEC. We also confirmed the previously reported association of the APOE/C1 locus and identified two novel loci (KLKB1 and CLSTN2) associated with CEC (Atherosclerosis 2023c).
Mitochondrial DNA
Hansi Weissensteiner, Lukas Forer, Florian Kronenberg, Sebastian Schönherr
Our group is strongly interested in analysing mitochondrial genomes (mtDNA) and related research. Over the past two years, our research has focused on improving mtDNA analysis tools and adapting novel sequencing approaches for large-scale analysis. Furthermore, we have conducted extensive research to gain a better understanding of the genetic factors contributing to the mtDNA abundance in one of the largest quantitative polymerase chain reaction (qPCR) studies. This involved further investigating the association of mtDNA copy numbers in patients with age-related macular degeneration.
- Haplogrep 3
In 2023 we published Haplogrep 3 (Nucleic Acids Res 2023), a significant upgrade to a widely used tool for classifying human mitochondrial DNA (mtDNA) haplogroups. Having been cited over 1,500 times, Haplogrep 3 now offers a range of enhanced features for large-scale genomic analysis. These include variant annotation from databases such as GnomAD, flexible phylogenetic tree management, an interactive web interface, improved FASTA classification and pre-classification quality control for VCF files. Our aim is to provide a more powerful platform for investigating mitochondrial DNA variation in various research fields, particularly medical, forensic and evolutionary genetics. Haplogrep is available as both a web service and for local installation to guarantee accessibility to a large number of users. More details can be found at https://haplogrep.i-med.ac.at.
- mtDNA-Server 2
In 2024 we updated our mtDNA sequencing analysis service in the cloud and described the major improvements to the pipeline (Nucleic Acids Res 2024a). In short, mtDNA-Server 2 enables users to analyse mtDNA sequencing data ranging from next-generation to long-read directly in the cloud, eliminating the need for hardware installation and maintenance. We have now changed the underlying architecture of our pipeline and improved the validation steps to enhance quality control. We have also included GATK Mutect2 for calling short insertions and deletions, integrated the contamination check from our Haplocheck tool and taken advantage of Haplogrep 3 by providing interactive variant reports. The pipeline can also be run on local hardware; more details can be found on our Mitoverse platform: https://mitoverse.i-med.ac.at/.
- Genome-wide association study on mtDNA copy number
We conducted a meta-GWAS (n=16,130) including data from the CHRIS study, GCKD and the AugUR study to identify genetic loci associated with qPCR-measured mitochondrial DNA copy number (mtDNA-CN). We discovered two genome-wide significant autosomal loci at HBS1L and GSDMA. Additionally, we compared qPCR-derived mtDNA-CN with microarray-based estimates and found strong concordance for most previously reported single nucleotide polymorphisms (SNPs). Our results suggest that the mitochondrial genome itself plays only a minor role in mtDNA-CN regulation (Sci. Rep. 2024).
- mtDNA and age-related macular degeneration (AMD)
We investigated the relationship between mitochondrial DNA copy number (mtDNA-CN) and age-related macular degeneration (AMD)in 2,262 elderly participants from the AugUR study. Using a quantitative polymerase chain reaction (qPCR) approach with plasmid normalisation, we found significantly lower mtDNA-CN in participants with early (n=453) and late (n=170) AMD compared to AMD-free individuals (n=1,630). Regression analyses revealed that a decrease of one standard deviation in mtDNA-CN increased the risk of late AMD by 24%. The strongest association was observed in geographic atrophy, where the risk was 76% higher. These results imply that mtDNA-CN in blood could be a potential AMD biomarker, offering a more accessible alternative to retinal mtDNA-CN measurements (Int. J. Mol. Sci. 2023).
Computational Genomics, Genome Informatics and Statistical Genetics
Sebastian Schönherr, Lukas Forer, Claudia Lamina, Florian Kronenberg, Hansi Weissensteiner, Johanna Schachtl-Rieß, Silvia Di Maio
Developing novel algorithms and computational tools to explore the genetics of various phenotypes.
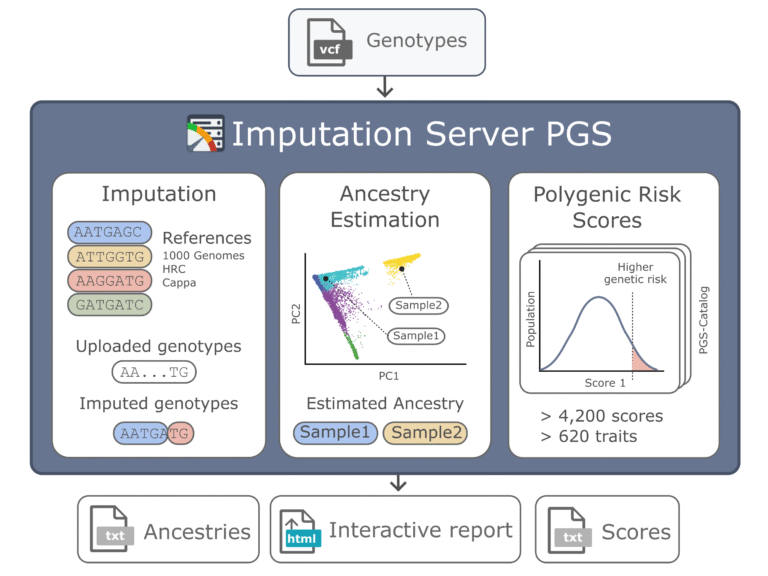
- Imputation Server
We have introduced the Imputation Server PGS (Nucleic Acids Research 2024b), which is an extension of the Michigan Imputation Server that was developed by our institute.
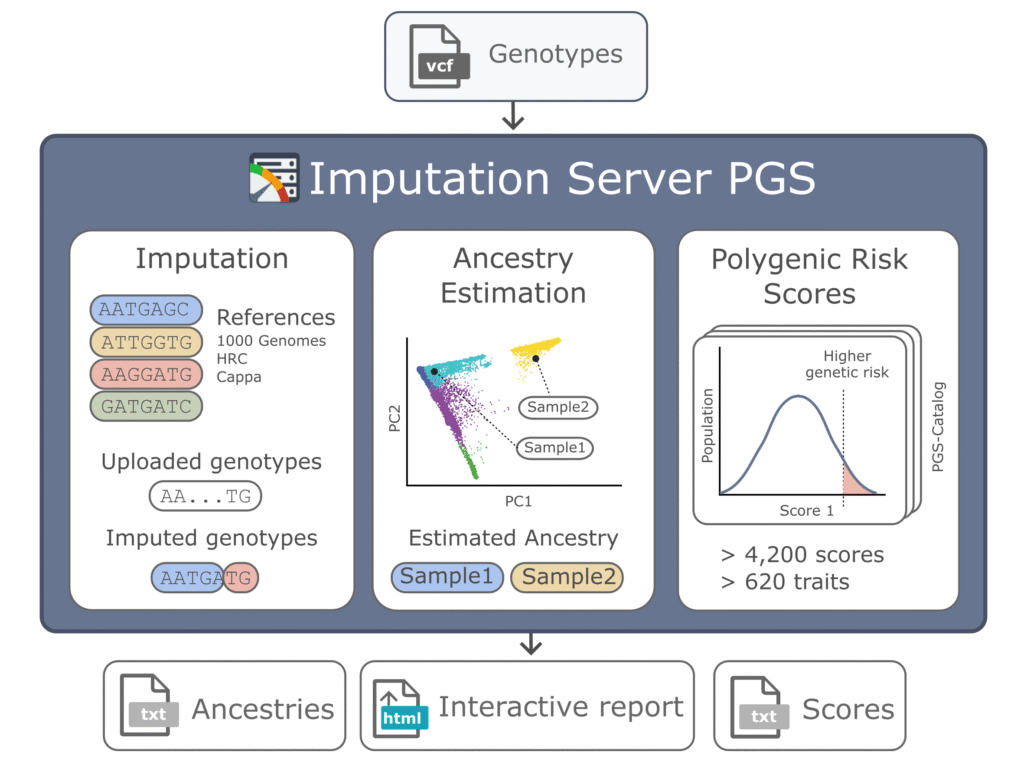
It automates and standardises polygenic scores (PGS) using high-quality genotypes and multi-ethnic haplotype panels. To date, over 13,000 users have imputed more than 120 million genomes, generating over 1 petabyte of results. The service supports over 5,000 published PGSs and includes quality control features such as ancestry estimation and an interactive report to facilitate efficient score comparison. The service is freely available at https://imputationserver.sph.umich.edu.
- Genetic Risk scores
Polygenic Risk Scores (PRS) are powerful tools for predicting an individual’s genetic predisposition to various traits and diseases. This approach has the potential to enable early risk prediction and personalised medicine. Over the past two years, our institute has developed high-throughput methods for efficiently calculating polygenic scores, allowing large-scale assessments to be conducted across diverse populations. We have also investigated and evaluated the potential of PGS for understanding complex traits and diseases. For example, in a population-based study from Southern Germany (Sci.Rep. 2023), we identified a strong and independent association between a genome-wide polygenic score and a family history score with the risk of prevalent and incident Type 2 Diabetes (T2D). This suggests that including a PGS in family history assessments could improve T2D prediction.
- A pipeline for reproducible biobank-scale GWAS
GWAS are a vital tool for identifying new relationships between genotypes and phenotypes. However, increasing sample sizes present researchers with computational challenges. To enable them to focus on data interpretation, we created nf-gwas, a Nextflow pipeline to run biobank-scale GWAS. The pipeline automatically performs pre- and post-processing steps, integrating regression modelling from the REGENIE package. NF-GWAS includes extensive reporting functionality and has been tested using nf-test, which is a crucial requirement in clinical and pharmaceutical settings. Furthermore, we have validated the pipeline against published GWAS datasets and benchmarked it on high-performance computing and cloud infrastructures in order to provide cost estimations for end users. In conclusion, nf-gwas is a highly parallelised, scalable and well-tested Nextflow pipeline for performing GWAS analyses in a reproducible manner (NAR Genom Bioinform, 2024).
The structural basis for Psychotropic Drug Biosynthesis
Sebastiaan Werten, Bernhard Rupp
The structural biology team supports interdisciplinary projects by providing detailed three-dimensional molecular structures of disease-related proteins. This is achieved through experimental methods such as X-ray diffraction, cryo-EM and in silico modelling. The team focuses on novel psychoplastogens containing 4-hydroxy-tryptamine with therapeutic potential for treating therapy-resistant depression and related CNS disorders.
The structural basis of Psilocybin Biosynthesis
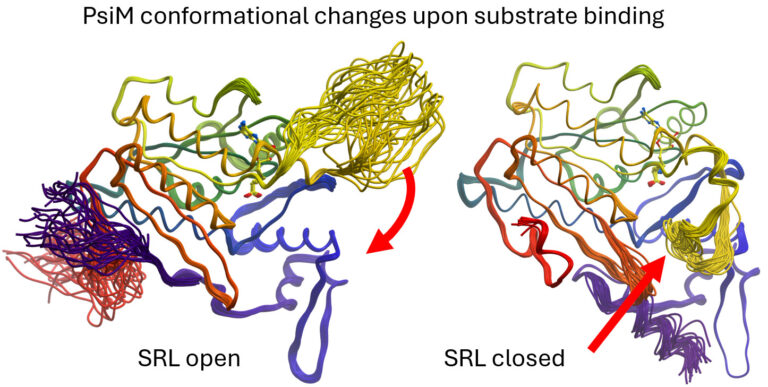
As the lead investigators of an international DACH project funded by the Austrian Science Fund (FWF), we are studying the biosynthesis of psilocybin, the active psychotropic compound found in magic mushrooms of the Psilocybe genus, using X-ray crystallography. As early as 2022, we identified psiD as a non-canonical, PLP-independent PSD-II decarboxylase. In 2024, we deciphered the intricate two-stage methylation process by PsiM, the final step converting norbaeocystin to baeocystin and finally Psilocybin (Nat.Commun. 2024).
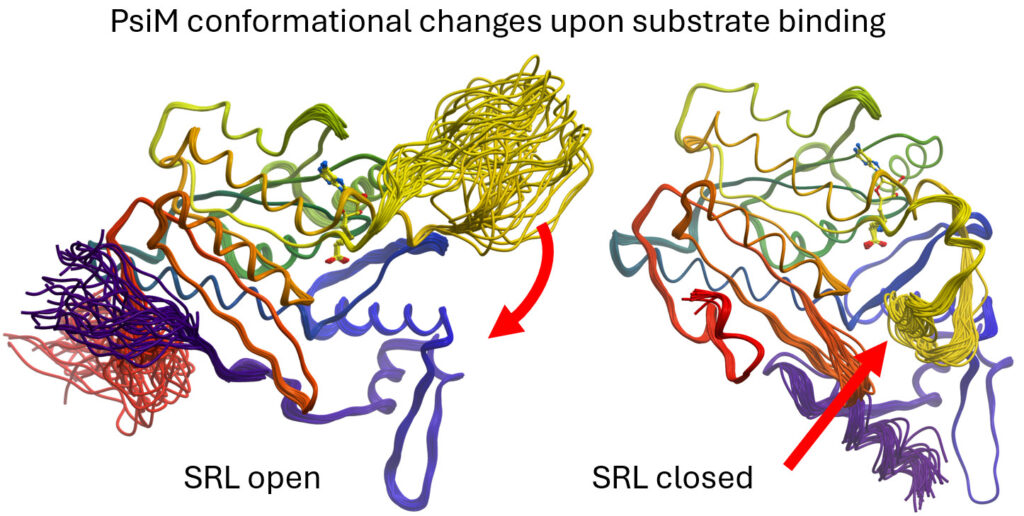
We then conducted a study that established the dynamic conformational changes of the substrate recognition loop of PsiM (J.Appl.Cryst. 2025). These structural insights enable the design of biosynthetic pathways for novel hydroxy tryptamine psychoplastogens with therapeutic potential for treating therapy-resistant depression and related CNS disorders.
Pictures
Selected Publications
Kollerits B, Kotsis F, Schneider MP, Schultheiss UT, Weissensteiner H, Schönherr S, Forer L, Meiselbach H, Wanner C, Eckardt KU, Dieplinger H, Kronenberg F, GCKD Investigators: Association of serum afamin concentrations with kidney failure in patients with CKD: Findings from the German CKD cohort study. Am. J. Kidney Dis. 85:432, 2025. PMID: 39743167
Kronenberg F, Mora S, Stroes ESG, Ference BA, Arsenault BJ, Berglund L, Dweck MR, Koschinsky ML, Lambert G, Mach F, McNeal CJ, Moriarty PM, Natarajan P, Nordestgaard BG, Parhofer KG, Virani SS, von Eckardstein A, Watts GF, Stock JK, Ray KK, Tokgözoğlu LS, Catapano AL: Frequent questions and responses on the 2022 lipoprotein(a) consensus statement of the European Atherosclerosis Society. Atherosclerosis 374:107-120, 2023a. PMID: 37188555
Kheirkhah A, Schachtl-Riess JF, Lamina C, Di Maio S, Koller A, Schönherr S, Coassin S, Forer L, Sekula P, Gieger C, Peters A, Köttgen A, Eckardt KU, Kronenberg F: Meta-GWAS on PCSK9 concentrations reveals associations of novel loci outside the PCSK9 locus in White populations. Atherosclerosis 386:117384, 2023b. PMID: 37989062
Kronenberg F, Bedlington N, Ademi Z, Geanta M, Silberzahn T, Rijken M, Kaal A, Harada-Shiba M, Chen Z, Thanassoulis G, Eliasen B, Eisele JL, Wiegman A, Ballantyne CM, Broome E, Calabro M, Corral P, Dol A, Donato LJ, Evans E, Funabashi S, Gouni-Berthold I, Ibarluzea IG, Johnson N, Lane J, Mora S, Nordestgaard BG, Pecin I, Kaal-Poppelaars R, Langlois MR, Ray KK, Rodenbach A, Santos RD, Stroes ESG, Tada H, Vrablik M, Winokur M, Yoshida M, Nicholls SJ, Daccord M: The Brussels International Declaration on Lipoprotein(a) Testing and Management. Atherosclerosis 406: 119218, 2025. PMID: 40340180
Schachtl-Riess JF, Schönherr S, Lamina C, Forer L, Coassin S, Streiter G, Kheirkhah A, Li Y, Meiselbach H, Di Maio S, Eckardt KU, Köttgen A, Kronenberg F, GCKD investigators: KLKB1 and CLSTN2 are associated with HDL-mediated cholesterol efflux capacity in a genome-wide association study. Atherosclerosis 368:1-11, 2023c. PMID: 36812656
Kollerits B, Gruber S, Steinbrenner I, Schwaiger JP, Weissensteiner H, Schönherr S, Forer L, Kotsis F, Schultheiss UT, Meiselbach H, Wanner C, Eckardt KU, Kronenberg F, GCKD Investigators: Apolipoprotein A-IV concentrations and cancer in a large cohort of chronic kidney disease patients: results from the GCKD study. BMC Cancer 24:320, 2024. PMID: 38454416
Noce D, Foco L, Orth-Höller D, König E, Barbieri G, Pietzner M, Ghasemi-Semeskandeh D, Coassin S, Fuchsberger C, Gögele M, Del Greco M F, De Grandi A, Summerer M, Wheeler E, Langenberg C, Lass-Flörl C, Pramstaller PP, Kronenberg F*, Würzner R*, Pattaro C*: Genetic determinants of complement activation in the general population. Cell Rep 43:113611, 2024. PMID: 38159276
Gollmann-Tepeköylü C, Graber M, Hirsch J, Mair S, Naschberger A, Pölzl L, Nägele F, Kirchmair E, Degenhart G, Demetz E, Hilbe R, Chen HY, Engert JC, Böhm A, Franz N, Lobenwein D, Lener D, Fuchs C, Weihs A, Töchterle S, Vogel GF, Schweiger V, Eder J, Pietschmann P, Seifert M, Kronenberg F, Coassin S, Blumer M, Hackl H, Meyer D, Feuchtner G, Kirchmair R, Troppmair J, Krane M, Weiss G, Tsimikas S, Thanassoulis G, Grimm M, Rupp B, Huber LA, Zhang SY, Casanova JL, Tancevski I, Holfeld J: Toll-like receptor 3 mediates aortic stenosis through a conserved mechanism of calcification. Circulation 147:1518-1533, 2023. PMID: 37013819
Dikaios I, Althaus H, Angles-Cano E, Ceglarek U, Coassin S, Cobbaert CM, Delatour V, Dieplinger B, Grimmler M, Hoofnagle AN, Kostner GM, Kronenberg F, Kuklenyik Z, Lyle AN, Prinzing U, Ruhaak LR, Scharnagl H, Vesper HW, Deprez L: Commutability assessment of candidate reference materials for lipoprotein(a) by comparison of a MS-based candidate reference measurement procedure with immunoassays. Clin. Chem. 69:262-272, 2023. PMID: 36644921
Ruhaak LR, Romijn FPHTM, Begcevic Brkovic I, Kuklenyik Z, Dittrich J, Ceglarek U, Hoofnagle AN, Althaus H, Angles-Cano E, Coassin S, Delatour V, Deprez L, Dikaios I, Kostner GM, Kronenberg F, Lyle A, Prinzing U, Vesper HW, Cobbaert CM: Development of an LC-MRM-MS-based candidate reference measurement procedure for standardization of serum apolipoprotein (a) tests. Clin. Chem. 69:251-261, 2023. PMID: 36644914
Kronenberg F: Lipoprotein(a): from Causality to Treatment. Curr Atheroscler Rep 26:75-82, 2024. PMID: 38252372
Mark PB, Carrero JJ, Matsushita K, Sang Y, Ballew SH, Grams ME, Coresh J, Surapaneni A, Brunskill NJ, Chalmers J, Chan L, Chang AR, Chinnadurai R, Chodick G, Cirillo M, de Zeeuw D, Evans M, Garg AX, Gutierrez OM, Heerspink HJL, Heine GH, Herrington WG, Ishigami J, Kronenberg F, Lee JY, Levin A, Major RW, Marks A, Nadkarni GN, Naimark DMJ, Nowak C, Rahman M, Sabanayagam C, Sarnak M, Sawhney S, Schneider MP, Shalev V, Shin JI, Siddiqui MK, Stempniewicz N, Sumida K, Valdivielso JM, van den Brand J, Yee-Moon Wang A, Wheeler DC, Zhang L, Visseren FLJ, Stengel B: Major cardiovascular events and subsequent risk of kidney failure with replacement therapy: a CKD Prognosis Consortium study. Eur. Heart J. 44:1157-1166, 2023. PMID: 36691956
Di Maio S, Zöscher P, Weissensteiner H, Forer L, Schachtl-Riess JF, Amstler S, Streiter G, Pfurtscheller C, Paulweber B, Kronenberg F, Coassin S, Schönherr S: Resolving intra-repeat variation in medically relevant VNTRs from short-read sequencing data using the cardiovascular risk gene LPA as a model. Genome Biol. 25:167, 2024. PMID: 38926899
Amstler S, Streiter G, Pfurtscheller C, Forer L, Di Maio S, Weissensteiner H, Paulweber B, Schönherr S, Kronenberg F, Coassin S: Nanopore sequencing with unique molecular identifiers enables accurate mutation analysis and haplotyping in the complex lipoprotein(a) KIV-2 VNTR. Genome Med. 16:117, 2024. PMID: 39380090
Koller A, Lamina C, Brandl C, Zimmermann ME, Stark KJ, Weissensteiner H, Würzner R, Heid IM, Kronenberg F: Systemic evidence for mitochondrial dysfunction in age-related macular degeneration as revealed by mtDNA copy number measurements in peripheral blood. Int. J. Mol. Sci. 24:16406, 2023. PMID: 38003595
Tschiderer L, Innerhofer H, Seekircher L, Waltle L, Richter L, Kimpel J, Lass-Flörl C, Forer L, Schönherr S, Larsen DA, Krammer F, Embacher-Aichhorn S, Tilg H, Weiss G, Allerberger F, Willeit P: Long-term effectiveness of an ultra-rapid rollout vaccination campaign with BNT162b2 on the incidence of SARS-CoV-2 infection. iScience 27:111117, 2024. PMID: 39555399
Mora S, Kronenberg F: Lipoprotein(a). JAMA 333: 1918-1919, 2025. PMID: 40272827
Tsimikas S, Yeang C, Kronenberg F: In search of an accurate measurement of LDL-C: Correction for Lp(a)-cholesterol to predict clinical outcomes. J. Am. Coll. Cardiol. 84:178-181, 2024. PMID: 38960511
Leitner L, Hudspeth J, Werten S, Rupp B: If you cannot see it, is it still there? Journal of Applied Crystallography 58:615, 2025. PMID:
Gruber I, Kollerits B, Forer L, Di Maio S, Schachtl-Riess JF, Kheirkhah A, Schönherr S, Schultheiss UT, Köttgen A, Eckardt KU, Coassin S, Lamina C, Kronenberg F: Lipoprotein(a) concentrations and cardiovascular disease in patients with chronic kidney disease: Results from the German Chronic Kidney Disease study. J. Intern. Med. 296:510-526, 2024. PMID: 39513193
Seekircher L, Bánki Z, Kimpel J, Rössler A, Schäfer H, Falkensammer B, Bante D, Forer L, Schönherr S, Shieldvacc-2 Study Group, Harthaller T, Sacher M, Ower C, Tschiderer L, Ulmer H, Krammer F, von Laer D, Borena W, Willeit P: Immune response after two doses of the BNT162b2 COVID-19 vaccine and risk of SARS-CoV-2 breakthrough infection in Tyrol, Austria: an open-label, observational phase 4 trial. Lancet Microbe 4:e612-e621, 2023. PMID: 37354911
Schönherr S*, Schachtl-Riess JF, Di Maio S*, Filosi M, Mark M, Lamina C, Fuchsberger C, Kronenberg F, Forer L: Performing highly parallelized and reproducible GWAS analysis on biobank-scale data. NAR Genom Bioinform 6:lqae015, 2024. PMID: 38327871
Hudspeth J, Rogge K, Dörner S, Müll M, Hoffmeister D, Rupp B, Werten S: Methyl transfer in psilocybin biosynthesis. Nat. Commun. 15:2709, 2024. PMID: 38548735
Schlosser P, Scherer N, Grundner-Culemann F, Monteiro-Martins S, Haug S, Steinbrenner I, Uluvar B, Wuttke M, Cheng Y, Ekici AB, Gyimesi G, Karoly ED, Kotsis F, Mielke J, Gomez MF, Yu B, Grams ME, Coresh J, Boerwinkle E, Köttgen M, Kronenberg F, Meiselbach H, Mohney RP, Akilesh S, GCKD Investigators, Schmidts M, Hediger MA, Schultheiss UT, Eckardt KU, Oefner PJ, Sekula P, Li Y, Köttgen A: Genetic studies of paired metabolomes reveal enzymatic and transport processes at the interface of plasma and urine. Nat. Genet. 55:995-1008, 2023. PMID: 37277652
Schönherr S, Weissensteiner H, Kronenberg F, Forer L: Haplogrep 3 – an interactive haplogroup classification and analysis platform. Nucleic Acids Res. 51:W263-W268, 2023a. PMID: 37070190
Weissensteiner H*, Forer L*, Kronenberg F, Schönherr S: mtDNA-Server 2: advancing mitochondrial DNA analysis through highly parallelized data processing and interactive analytics. Nucleic Acids Res. 52:W102-W107, 2024a. PMID: 38709886
Forer L, Taliun D, LeFaive J, Smith AV, Boughton AP, Coassin S, Lamina C, Kronenberg F, Fuchsberger C, Schönherr S: Imputation Server PGS: an automated approach to calculate polygenic risk scores on imputation servers. Nucleic Acids Res. 52:W70-W77, 2024b. PMID: 38709879
Koschinsky ML, Stroes ESG, Kronenberg F: Daring to dream: Targeting lipoprotein(a) as a causal and risk-enhancing factor. Pharmacol. Res. 194:106843, 2023. PMID: 37406784
Koller A, Filosi M, Weissensteiner H, Fazzini F, Gorski M, Pattaro C, Schönherr S, Forer L, Herold JM, Stark KJ, Döttelmayer P, Hicks AA, Pramstaller PP, Würzner R, Eckardt KU, Heid IM, Fuchsberger C, Lamina C, Kronenberg F: Nuclear and mitochondrial genetic variants associated with mitochondrial DNA copy number. Sci. Rep. 14:2083, 2024. PMID: 38267512
Duschek E, Forer L, Schönherr S, Gieger C, Peters A, Kronenberg F, Grallert H, Lamina C: A polygenic and family risk score are both independently associated with risk of type 2 diabetes in a population-based study. Sci. Rep. 13:4805, 2023. PMID: 36959271
Heilmann E, Costacurta F, Moghadasi SA, Ye C, Pavan M, Bassani D, Volland A, Ascher C, Weiss AKH, Bante D, Harris RS, Moro S, Rupp B, Martinez-Sobrido L, von Laer D: SARS-CoV-2 3CLpro mutations selected in a VSV-based system confer resistance to nirmatrelvir, ensitrelvir, and GC376. Sci. Transl. Med. 15:eabq7360, 2023. PMID: 36194133
* authors contributed equally
Selection of Funding
- “Haplotypenanalyse im LPA KIV-2 CNV und Imputation: Multi-ancestry LPA KIV-2 haplotype analysis and imputation”, FWF, Stefan Coassin
- “Cytokines – causal Lp(a) determinants?”, Austrian Atherosclerosis Society, Johanna F. Schachtl-Riess
- “Disentangling the mystery around free apo(a)”, Austrian Atherosclerosis Society, Adriana Koller
- “Epidemiological and genetic evidence for an influence of apolipoprotein A-IV on health and disease”, Tiroler Nachwuchsforscher*innenförderung, Adriana Koller
- “Decoding PCSK9: New Perspectives on the Mature and Furin-Cleaved Forms”, Wael Almahmeed Research Training Fellowship from the International Atherosclerosis Society, Azin Kheirkhah
- “Askimed-Cloud-Plattform zur Erhebung, Verwaltung und Analyse von Daten im Gesundheitssektor”, Lukas Forer, Sebastian Schönherr
Collaborations
- Gonçalo Abecasis, Center of Statistical Genetics, University of Michigan, Ann Arbor, USA
- KORA Studies: Institute of Epidemiology, Helmholtz Zentrum München, Neuherberg, Germany
- Kai-Uwe Eckardt, Department of Nephrology and Medical Intensive Care, Charité—Universitätsmedizin Berlin, Berlin, Germany
- Nicole Probst-Hensch, Swiss Tropical and Public Health Institute, Basel, Switzerland
- Anna Köttgen, Institute of Genetic Epidemiology, University of Freiburg, Freiburg, Germany
- Iris M. Heid, Department of Genetic Epidemiology, University of Regensburg, Regensburg, Germany
- CoLAUS Study: Peter Vollenweider / Pedro Marques-Vidal, Department of Medicine, Internal Medicine, Lausanne University Hospital (CHUV), Lausanne, Switzerland
- Young Finns Study (YFS), Terho Lehtimäki, Tampere University/ Olli Raitakari, University of Turku, Finland
- Dirk Hofmeister, Institute of Pharmacy, Friedrich Schiller University, Jena, Germany; Research Group Pharmaceutical Microbiology, Leibniz Institute of Natural Product Research and Infection Biology; Hans Knöll Institute; Jena, Germany