Innrain 80
6020 Innsbruck
Fax: +43 (0)512 9003 73960
Email: Andreas.Villunger@i-med.ac.at
Website: https://www.villungerlab.com/
Research year
Research Branch (ÖSTAT Classification)
301105, 301108, 301902, 301904
Keywords
Cell Death, DNA methylation, Haematopoiesis, Immunology, Inflammation, Non-coding RNAs, and Tumour Biology
Research Focus
Together with their co-workers, IDI investigators are exploring basic mechanisms of immune-cell development and function with an emphasis on cell death – cell-cycle crosstalk, innate and adaptive immunity, microRNAs and steroid hormones. We are also studying general principles of cellular transformation, focusing on the role of BCL2-regulated cell death and the p53 signalling network as a barrier against malignant disease. For additional details, please see https://www.villungerlab.com/
General Facts
The Institute is part of the Biocenter (https://biocenter.i-med.ac.at ) and hosts four research groups, run by Andreas Villunger and associate professors Joel Riley, Verena Labi and Sebastian Herzog. The local resources and infrastructure include ample modern lab space, preclinical models of malignant disease, immune-deficiency and autoimmunity, S2-tissue culture for lenti- or retroviral transduction of model cell lines, histology support, cell sorting (BD FACS_ARIAIII), multi-colour flow cytometry (BD FACS-LSR-Fortessa), live-cell imaging (IncuCyte) and a CRISPR/Cas9-based screening platform. The Center of Molecular Medicine (CeMM) of the Austrian Academy of Science in Vienna, where Andreas Villunger holds an adjunct PI position, offers rapid access to PLACEBO drug screening and the biomedical sequencing facility (BSF) for RNAseq studies (https://cemm.at).
All faculty members contribute to teaching and training of students enrolled in human medicine or life sciences at bachelor and master levels. A structured PhD programme secures a first-class training environment for the PhD students at our institute (https://phd-school.i-med.ac.at/). Scientific exchange is guaranteed at a range of levels, including regular group meetings, institute and department seminars and participation in international conferences.
Research
The Cell Death Signalling Lab
Andreas Villunger
The Cell Death Signalling lab is run by the Head of the Institute, Andreas Villunger, and is currently exploring the cross talk between the cell-death and the cell-cycle machineries. The focus lies on the signaling events that define thresholds for cell death or survival after mitotic errors. The group is considering two key questions: how are mitotic errors communicated to alert the immune system? and how is p53, a major tumour suppressor, engaged in this context, e.g. in response to centrosomal abnormalities? We have shown that PIDD1, sitting at the centrosome in healthy cells
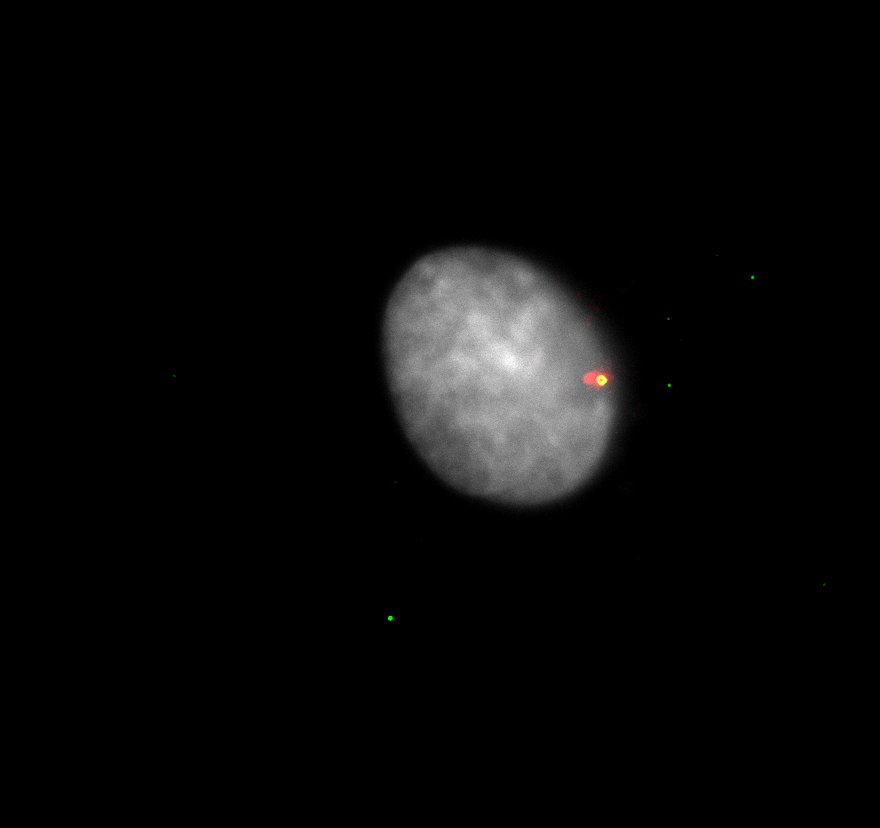
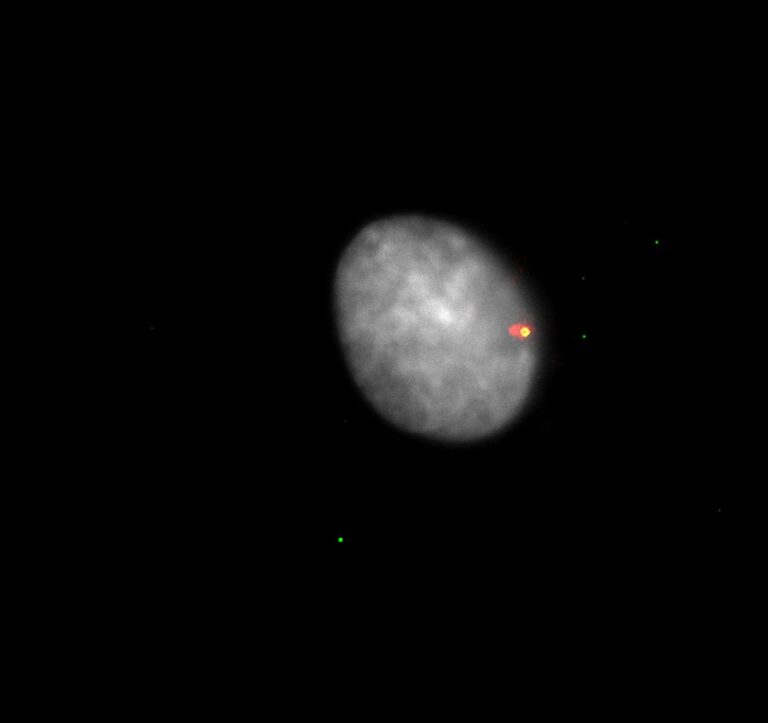
can trigger sterile inflammation when centrosomes accumulate.
The team is also investigating the consequences of centrosome amplification for tissue homeostasis. Overexpression of Polo-like Kinase 4 (PLK4) leads to centrosome amplification and engages the PIDDosome, which drives p53 activation and expression of the cell cycle regulator, p21.
It is believed that this process limits transformation but it may also be the desired outcome during normal organ development, as exemplified in the liver or heart.
The Cell Death & Inflammation Lab
Joel Riley
Immunogenicity of cell death
Cell death, in particular programmed cell death modalities such as apoptosis, have long been considered to be immune-silent, as they do not trigger any immune response. However, it is becoming clear that under some circumstances apoptosis can be highly pro-inflammatory. We have shown that when caspases are inhibited (genetically or pharmacologically) apoptosis triggers a potent immune response. This is in part due to the release of mitochondrial DNA into the cytoplasm, where it is sensed by innate immune sensors such as cGAS-STING, initiating a potent type I interferon response.
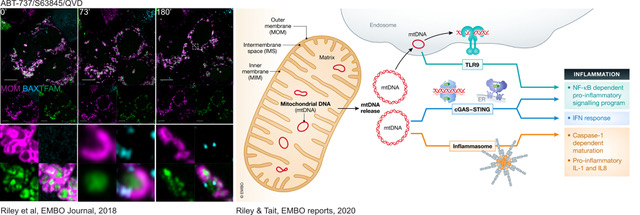
The consequences of mtDNA release during cell death are under active investigation and our work and that of others suggests that they could have a huge impact on the pathology of various diseases. We are currently trying to understand how mitochondria become hubs of inflammation during cell death.
Oncogenic cell death signalling
The ability to evade cell death is a hallmark of cancer. However, we have shown that low-level activation of the apoptotic machinery can be oncogenic, rather than anti-tumour. Using super-resolution imaging, we have demonstrated that fragmented, dysfunctional mitochondria accumulate pro-apoptotic proteins on their membranes, allowing a limited, but detectable, activation of caspases. Importantly, this level of caspase activation is not sufficient to cause cell death but does cause DNA damage, which can be oncogenic.
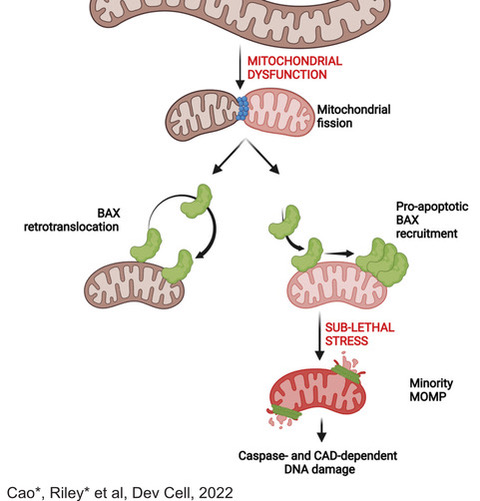
Current research in the lab is aimed at understanding how this process contributes to oncogenesis, particularly in B-cell lymphomagenesis, and how it can be harnessed for therapeutic gain.
The Experimental Immunology Lab
Verena Labi
In the Experimental Immunology lab run by the Deputy Head of the Institute, Verena Labi, we are studying molecular control mechanisms of cell fitness and fate decisions. By means of genetic cell -culture and mouse models we are aiming to understand normal cell differentiation in comparison to pathological conditions.
We have previously shown that the miR-17-92 microRNAs prevent cell death in the developing lung by repressing the pro-apoptotic BIM protein. Following this line of research, we have established an in vitro lung organoid culture system to test whether this molecular mechanism, vital for proper lung development, fosters cancer in the adult lung.
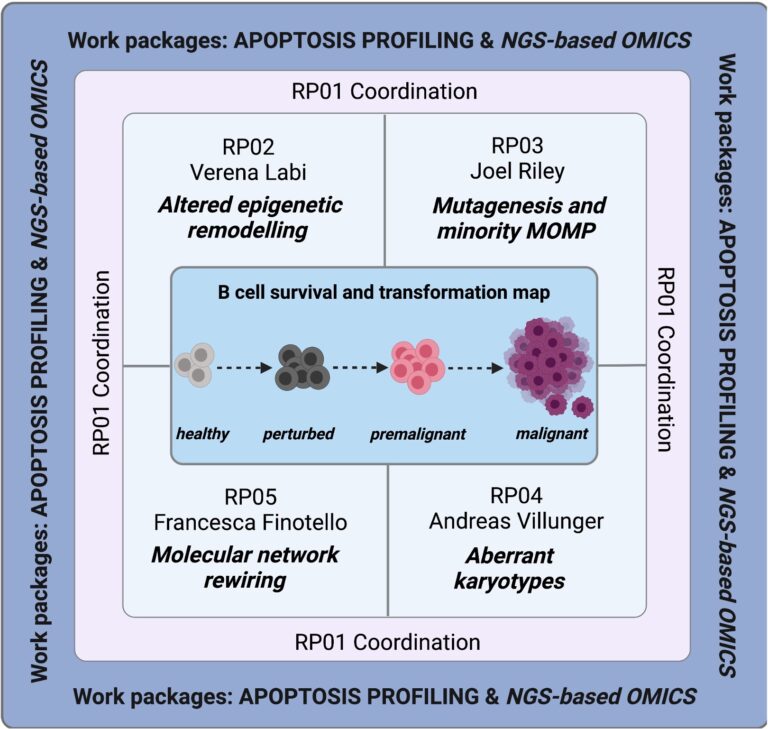
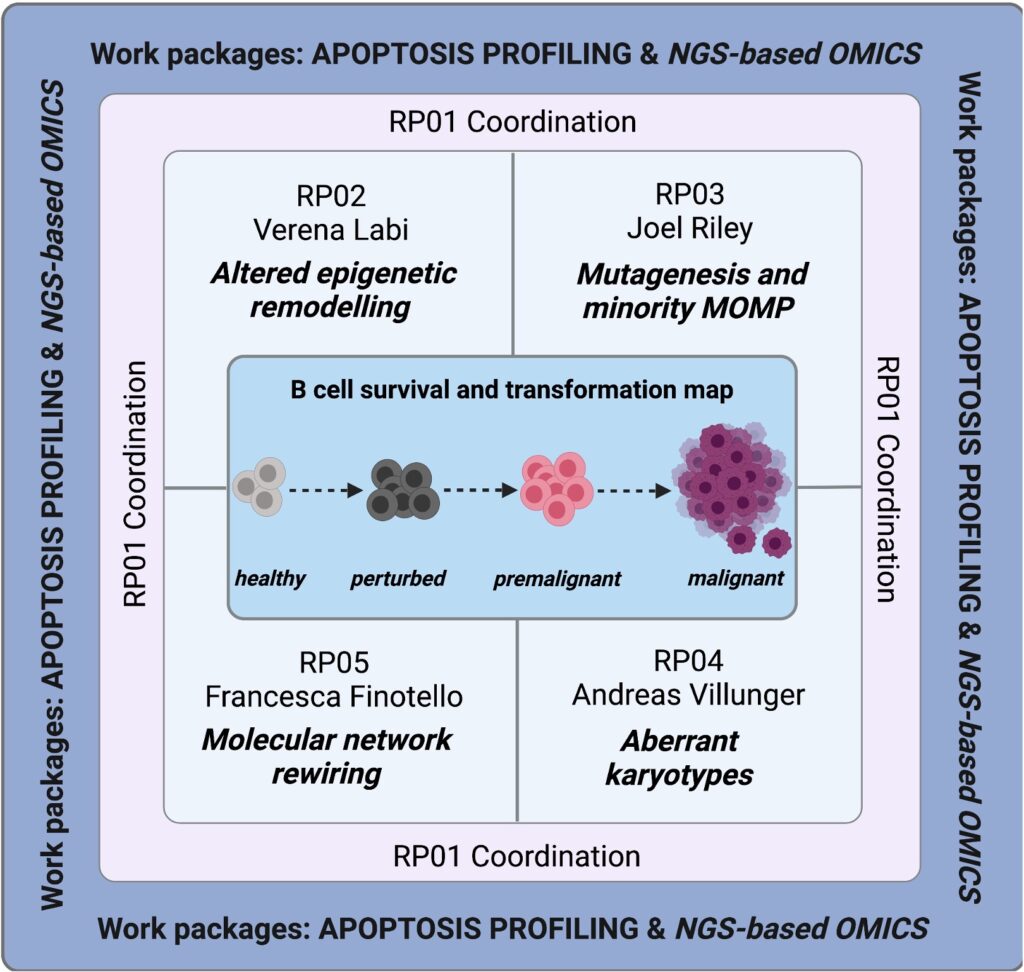
A second line of research is based on our finding that the epigenetic TET enzymes serve to establish and adapt transcriptional identity programmes during B-cell differentiation and activation. Notably, the TET enzymes ensure optimal humoral immunity while preventing pathologies such as immunodeficiency, autoimmunity and lymphomagenesis. We are trying to understand the pathologies associated with the loss of TET enzymes in B cells. We are also collaborating with A. Villunger and J. Riley from our Institute and with Francesca Finotello from the UIBK to study the molecular and cellular mechanisms by which TET2 suppresses tumours in B cells. We hope to develop a road map to the molecular rewiring of cell-death networks in B-cell transformation.
The Molecular Gene Regulation Lab
Sebastian Herzog
The non-protein coding part of the human genome comprises about 98 % of the ~3·109 DNA bases. It was initially considered to be “junk” DNA but it is extensively transcribed and gives rise to numerous non-coding RNAs including miRNAs, small non-coding RNAs with a critical role in post-transcriptional gene regulation. Our objective is to understand how microRNAs regulate gene expression on the molecular level and to what extend this regulation impacts cellular function.
MicroRNAs in B lymphocytes
We are aiming to decipher how miRNAs affect the sensitive processes that control B-cell development and activation that together ensure the establishment of a competent humoral immune system. We are approaching the topic by means of a two-pronged strategy, using gain-of-function and loss-of-function experiments and in vivo models to study transforming and physiologically relevant miRNAs.
MiRNA biogenesis
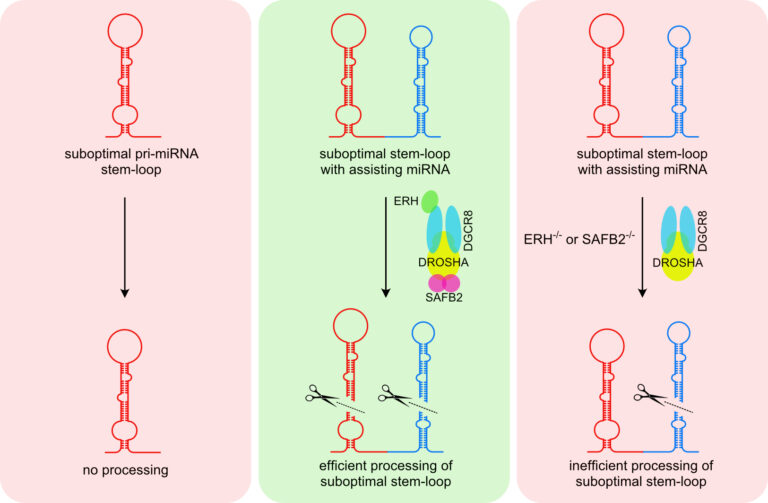
In canonical miRNA biogenesis, miRNA genes give rise to long primary transcripts (pri-miRNAs) characterized by one or several stem-loop structures in which the mature miRNAs are embedded. The stem-loop structures are recognized and cleaved by the nuclear Microprocessor, composed of the RNase DROSHA and its co-factor DGCR8. While most pri-miRNA are well defined by structural and sequence features, we do not fully understand how cells can discriminate miRNA-like stem-loop structures from authentic primary miRNAs or how accessory proteins beyond the core components shape the mature miRNA transcriptome.
We have recently characterized a novel mechanism called cluster assistance
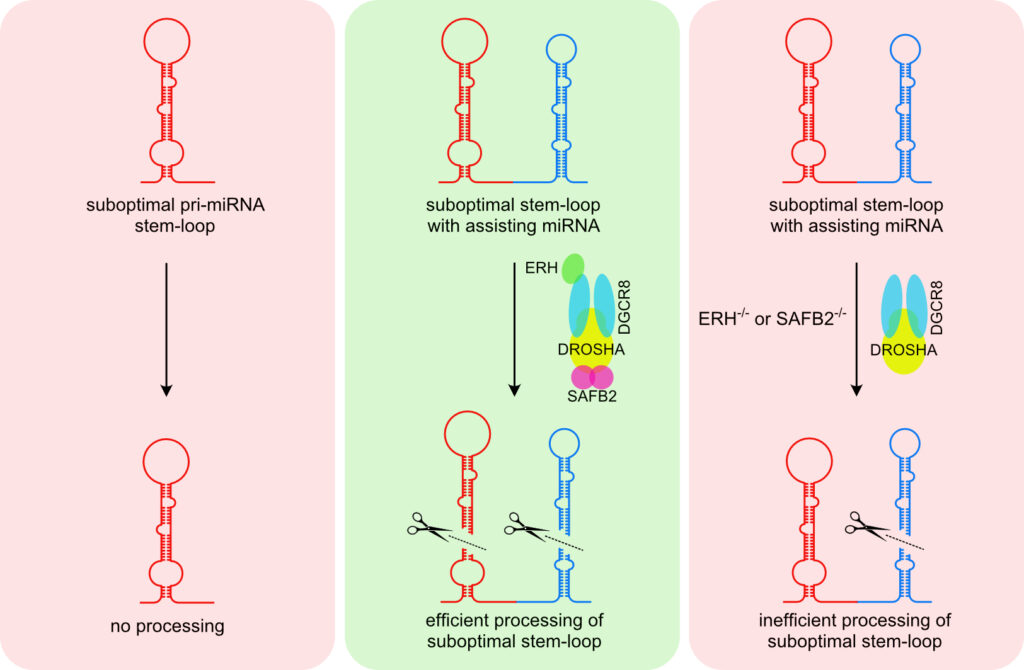
that significantly broadens the Microprocessor substrate specificity. A bona fide pri-miRNA stem-loop structure functions as a cis-regulatory element that can license or enhance the processing of neighbouring suboptimal pri-miRNAs within polycistronic clusters. However, we are far from understanding this phenomenon and are current aiming to unravel the mechanisms of cluster assistance and miRNA biogenesis in general.
Pictures
Selected Publications
Rizzotto D, Vigorito V, Rieder P, Gallob F, Moretta GM, Soratroi C, Riley JS, Bellutti F, Veli SL, Mattivi A, Lohmüller M, Herzog S, Bornhauser BC, Jacotot ED, Villunger A, Fava LL. Caspase-2 kills cells with extra centrosomes. Sci Adv. 2024 Nov;10(44):eado6607. doi: 10.1126/sciadv.ado6607. Epub 2024 Oct 30. PMID: 39475598; PMCID: PMC11524169.
Schapfl MA, LoMastro GM, Braun VZ, Hirai M, Levine MS, Kiermaier E, Labi V, Holland AJ, Villunger A. Centrioles are frequently amplified in early B cell development but dispensable for humoral immunity. Nat Commun. 2024 Oct 15;15(1):8890. doi: 10.1038/s41467-024-53222-4. PMID: 39406735; PMCID: PMC11480410.
Kiermaier E, Stötzel I, Schapfl MA, Villunger A. Amplified centrosomes-more than just a threat. EMBO Rep. 2024 Oct;25(10):4153-4167. doi: 10.1038/s44319-024-00260-0. Epub 2024 Sep 16. PMID: 39285247; PMCID: PMC11467336.
Karbon G, Schuler F, Braun VZ, Eichin F, Haschka M, Drach M, Sotillo R, Geley S, Spierings DC, Tijhuis AE, Foijer F, Villunger A. Chronic spindle assembly checkpoint activation causes myelosuppression and gastrointestinal atrophy. EMBO Rep. 2024 Jun;25(6):2743-2772. doi: 10.1038/s44319-024-00160-3. Epub 2024 May 28. PMID: 38806674; PMCID: PMC11169569.
Braun VZ, Karbon G, Schuler F, Schapfl MA, Weiss JG, Petermann PY, Spierings DCJ, Tijhuis AE, Foijer F, Labi V, Villunger A. Extra centrosomes delay DNA damage-driven tumorigenesis. Sci Adv. 2024 Mar 29;10(13):eadk0564. doi: 10.1126/sciadv.adk0564. Epub 2024 Mar 29. PMID: 38552015; PMCID: PMC10980279.
Vringer E, Heilig R, Riley JS, Black A, Cloix C, Skalka G, Montes-Gómez AE, Aguado A, Lilla S, Walczak H, Gyrd-Hansen M, Murphy DJ, Huang DT, Zanivan S, Tait SW. Mitochondrial outer membrane integrity regulates a ubiquitin-dependent and NF-κB-mediated inflammatory response. EMBO J. 2024 Mar;43(6):904-930. doi: 10.1038/s44318-024-00044-1. Epub 2024 Feb 9. PMID: 38337057; PMCID: PMC10943237.
Victorelli S, Salmonowicz H, Chapman J, Martini H, Vizioli MG, Riley JS, Cloix C, Hall-Younger E, Machado Espindola-Netto J, Jurk D, Lagnado AB, Sales Gomez L, Farr JN, Saul D, Reed R, Kelly G, Eppard M, Greaves LC, Dou Z, Pirius N, Szczepanowska K, Porritt RA, Huang H, Huang TY, Mann DA, Masuda CA, Khosla S, Dai H, Kaufmann SH, Zacharioudakis E, Gavathiotis E, LeBrasseur NK, Lei X, Sainz AG, Korolchuk VI, Adams PD, Shadel GS, Tait SWG, Passos JF. Apoptotic stress causes mtDNA release during senescence and drives the SASP. Nature. 2023 Oct;622(7983):627-636. doi: 10.1038/s41586-023-06621-4. Epub 2023 Oct 11. Erratum in: Nature. 2024 Jan;625(7995):E15. doi: 10.1038/s41586-023-07002-7. PMID: 37821702; PMCID: PMC10584674.
Bock FJ, Riley JS. When cell death goes wrong: inflammatory outcomes of failed apoptosis and mitotic cell death. Cell Death Differ. 2023 Feb;30(2):293-303. doi: 10.1038/s41418-022-01082-0. Epub 2022 Nov 14. PMID: 36376381; PMCID: PMC9661468.
Riley JS, Bock FJ. Voices from beyond the grave: The impact of apoptosis on the microenvironment. Biochim Biophys Acta Mol Cell Res. 2022 Nov;1869(11):119341. doi: 10.1016/j.bbamcr.2022.119341. Epub 2022 Aug 17. PMID: 35987283.
Cao K*, Riley JS*, Heilig R, Montes-Gómez AE, Vringer E, Berthenet K, Cloix C, Elmasry Y, Spiller DG, Ichim G, Campbell KJ, Gilmore AP, Tait SWG. Mitochondrial dynamics regulate genome stability via control of caspase-dependent DNA damage. Dev Cell. 2022 May 23;57(10):1211-1225.e6. doi: 10.1016/j.devcel.2022.03.019. Epub 2022 Apr 20. PMID: 35447090; PMCID: PMC9616799.
Weiss JG, Gallob F, Rieder P, Villunger A. Apoptosis as a Barrier against CIN and Aneuploidy. Cancers (Basel). 2022 Dec 21;15(1):30. doi: 10.3390/cancers15010030. PMID: 36612027; PMCID: PMC9817872.
Kim JY, Wang LQ, Sladky VC, Oh TG, Liu J, Trinh K, Eichin F, Downes M, Hosseini M, Jacotot ED, Evans RM, Villunger A, Karin M. PIDDosome-SCAP crosstalk controls high-fructose-diet-dependent transition from simple steatosis to steatohepatitis. Cell Metab. 2022 Oct 4;34(10):1548-1560.e6. doi: 10.1016/j.cmet.2022.08.005. Epub 2022 Aug 29. PMID: 36041455; PMCID: PMC9547947.
Lohmüller M, Roeck BF, Szabo TG, Schapfl MA, Pegka F, Herzog S, Villunger A, Schuler F. The SKP2-p27 axis defines susceptibility to cell death upon CHK1 inhibition. Mol Oncol. 2022 Aug;16(15):2771-2787. doi: 10.1002/1878-0261.13264. Epub 2022 Jul 7. PMID: 35673965; PMCID: PMC9348596.
Weiler ES, Szabo TG, Garcia-Carpio I, Villunger A. PIDD1 in cell cycle control, sterile inflammation and cell death. Biochem Soc Trans. 2022 Apr 29;50(2):813-824. doi: 10.1042/BST20211186. PMID: 35343572; PMCID: PMC9162469.
Cao K*, Riley JS*, Heilig R, Montes-Gomez, Vringer E, Berthenet K, Cloix C, Elmasry Y, Spiller DG, Ichim G, Campbell KJ, Gilmore AP, Tait SW. Mitochondrial dynamics regulate genome stability via control of caspase-dependent DNA damage. Dev Cell. 2022. May 23;57(10): 1211-1225. PMID 3544090
Hutter K, Rülicke T, Szabo TG, Andersen L, Villunger A, Herzog S. The miR-15a/16-1 and miR-15b/16-2 clusters regulate early B cell development by limiting IL-7 receptor expression. Front. Immunol. 2022 13:967914. doi: 10.3389/fimmu.2022.967914. PMID: 36110849. PMCID: PMC9469637.
Hutter K, Lindner SE, Kurschat C, Rülicke T, Villunger A and Herzog S. The miR-26 family regulates early B cell development and transformation. The Life Sci Alliance 2022 Apr 22;5(8):e202101303; doi: 10.26508/lsa.202101303. PMID: 35459737. PMCID: PMC9034462.]
Hagen M, Chakraborty T, Olson WJ, Heitz M, Hermann-Kleiter N, Kimpel J, Jenewein B, Pertoll J, Labi V, Rajewsky K, Derudder E. miR-142 favors naïve B cell residence in peripheral lymph nodes. Front Immunol. 2022 Nov 10;13:847415. doi: 10.3389/fimmu.2022.847415. eCollection 2022. PMID: 36439112
Erlacher M, Labi V. MCL-1 and BCL-XL: blood brothers. Blood. 2021 Apr 8;137(14):1850-1851. doi: 10.1182/blood.2020010569. PMID: 33830190
Hutter K, Lohmüller M, Jukic A, Eichin F, Avci S, Labi V, Szabo TG, Hoser SM, Hüttenhofer A, Villunger A, Herzog S. SAFB2 Enables the Processing of Suboptimal Stem-Loop Structures in Clustered Primary miRNA Transcripts. Mol Cell. 2020 Jun 4;78(5):876-889.e6; doi: 10.1016/j.molcel.2020.05.011. PMID: 32502422.
Selection of Funding
European Research Council: ERC_AdG – POLICE
FWF: Research Group “BEAT-IT” – BCL-2 Network Adaptations in B Cell Transformation
FWF/GCAR: Cyanobacterial Metabolites for Cancer Therapy
FWF: Research Group “BEAT-IT” – BCL-2 Network Adaptations in B Cell Transformation
TWF: Using advanced microscopy to understand cell death
FWF: Coordinator Research Group “BEAT-IT” – BCL-2 Network Adaptations in B Cell Transformation
FWF: Ferroptosis als Barriere gegen Blutkrebs; Grant-DOI:10.55776/P37290
FWF: Entscheidungen über Leben und Tod; Grant-DOI: 10.55776/I6642
FWF: Die Rolle von tRNA Thiolierung bei menschlichen Erkrankungen; Grant-DOI: 10.55776/PIN1183123
Collaborations
Alain de Bruin, Utrecht University, The Netherlands
Floris Foijer, ERIBA, Groningen, The Netherlands
Ana Garcia-Saez, University of Cologne, Germany
Luca Fava, CIBIO, Trento, Italy
Klaus Rajewsky, Max-Delbrück Center for Molecular Medicine, Berlin, Germany
Mir-Farzin Mashreghi, DRFZ, Berlin, Germany
Miriam Erlacher, Univ. Hospital, Freiburg, Germany
Gabriel Ichim, University of Lyon, France
Petr Broz, University of Lausanne, Switzerland
Yunsun Nam, University of Texas – Southwestern Medical Center, USA