Innrain 80-82
6020 Innsbruck
Email: David.Teis@i-med.ac.at
Website: https://biochem.i-med.ac.at/
Research year
Research Branch (ÖSTAT Classification)
106002, 106023, 106041, 106052
Keywords
Lipids, Membranes, metabolism, Molecular biochemistry, Proteostasis, and Signaling
Research Focus
Our Institute is aiming at a molecular understanding of how cells maintain a proper composition of their membrane proteins and lipids and how the processes contribute to human health. We are identifying molecular mechanisms for membrane quality control and membrane repair and characterizing how they are linked to lipid metabolism and how they are employed in cell-cell interactions (e.g. CAR T vs leukemia).
General Facts
The Institute of Molecular Biochemistry (IMB) is part of the Biocenter. Our research groups are aiming at a better understanding of the molecular mechanisms that build, maintain and repair cellular membranes. We are characterizing how the processes integrate with cellular lipid metabolism and how they are used to control cell–cell interactions. Defects lead to cellular damage that can result in human disease. We use a range of model organisms for genetic studies of membrane quality control at the molecular level, in combination with biochemical and cell biological approaches.
The Institute provides an international and dynamic environment for curiosity-driven students (MSc and PhD students) and postdoctoral fellows, with access to modern core facilities. Our PhD students are enrolled in the Molecular and Cellular Basis of Diseases (MCBD) PhD programme and we coordinate the “Cellular Basis of Diseases” doc.funds programme, which is funded by the FWF (DOC82). We are partners in two FWF-funded research groups on ‘Organelle proteostasis in cellular quiescence and growth’ (FG20) and ‘Damage & repair of membrane lipids in health and diseases’ (FG15).
Research
The Teis laboratory: MEMBRANE PROTEOSTASIS AND SIGNALING
The function of cells depends on the integrity of their membranes and organelles. To maintain membrane integrity, selective protein degradation machineries (cellular quality-control networks) detect and degrade mis-folded or orphaned proteins. This remarkable task maintains membrane protein homeostasis (proteostasis). Chronic defects in membrane proteostasis causes loss of organelle integrity and cell injuries that are central to the pathophysiologies of many human diseases, including cancer, autoimmunity, diabetes, obesity and neurodegeneration. The issue of how selective protein degradation pathways function and cooperate with membrane repair machineries is thus highly relevant to human health.
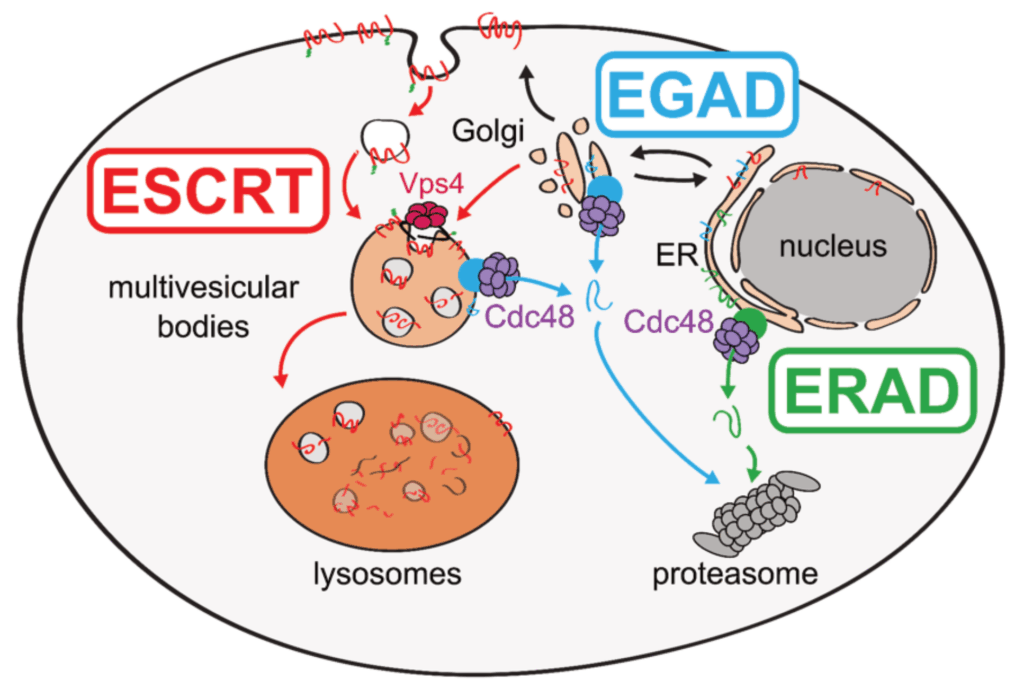
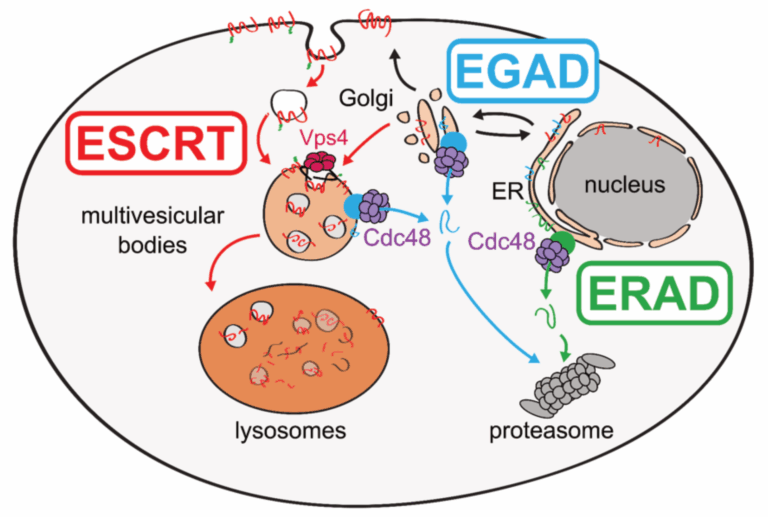
The Teis lab is studying the mechanisms that govern organelle proteostasis and how they integrate with cellular lipid metabolism, combining genetic approaches in yeasts and human cells with quantitative biochemical and imaging approaches. The work has (i) identified a membrane protein degradation pathway (EGAD) that selectively degrades orphaned membrane proteins at Golgi and endosomes to regulate TORC2-controlled lipid homeostasis (Weyer Y., et al., Nat. Comms, 2024; Schmidt O., et al., EMBO J., 2019); (ii) shown how metabolic signaling enlists ubiquitin ligase complex to degrade nutrient transporters as cells enter and exit quiescence (Ivashov V., et al., eLife, 2020; Kahlhofer J., et al., BioRxiv) and (iii) uncovered roles of the ESCRT machinery in regulating chromosome interactions with the nuclear envelope (Pieper G., et al., Dev. Cell, 2020) and in protecting the plasma membrane (Schmidt O., et al., JBC, 2020).
The Watschinger laboratory: ETHER LIPID METABOLISM
Ether lipids are important for physiological processes such as fine-structuring of the brain, protection of the eye from cataracts, sperm maturation and signaling. In recent years, it has become clear that conditions such as Alzheimer’s, Parkinson’s, chronic obstructive pulmonary disease and recently Covid-19 are linked to ether lipids, although the pathophysiological mechanisms are not clear.
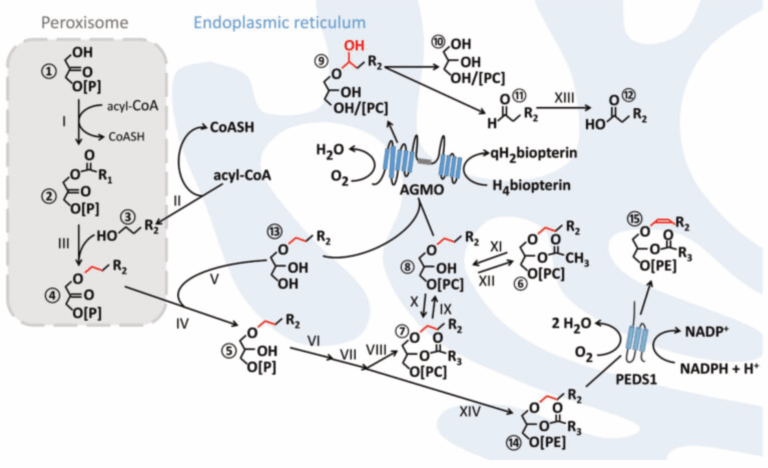
The Watschinger laboratory is tackling the challenge of unravelling the molecular mechanisms of ether lipid metabolism to increase our understanding of the processes that ultimately lead to disease in humans, potentially revealing novel targets for treatment in the future. Our focus lies on enzymes involved in ether lipid metabolism, e.g. alkylglycerol monooxyenase (AGMO) and plasmanylethanolamine desaturase (PEDS1), whose genes we have identified. While AGMO is an ether lipid-degrading enzyme, PEDS1 introduces the crucial vinyl ether double bond in plasmalogens. Key to the identification of the genes was the establishment of sensitive HPLC micro-assays based on a pyrene-labelled substrate. In a similar manner, we have recently established an assay for enzymes that degrade plasmalogens.
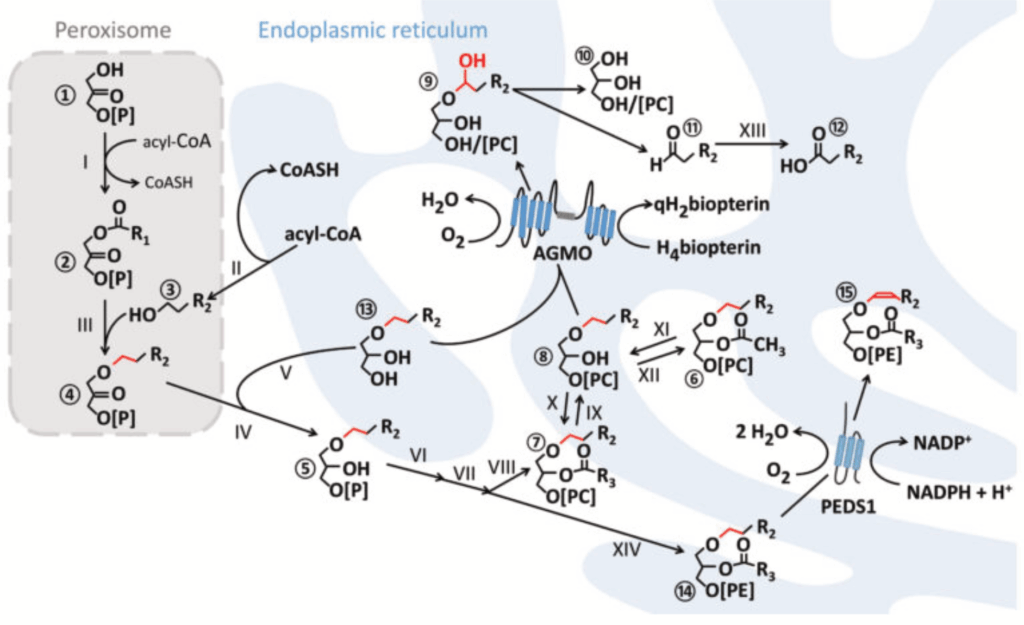
As structural information is now available, the genetic understanding and our sensitive assays are enabling us to investigate the (patho-) physiology of ether lipid metabolism in the various model systems we have established in our lab. By describing the physiological roles of ether lipids and their enzymes and with an optimal methodological toolkit, we will be able to provide experimental evidence for suspected genotype-phenotype correlations in humans.
The Müller laboratory: ENGINEERING IMMUNE CELLS FOR CANCER IMMUNOTHERAPY (together with Internal Medicine V; Dominik Wolf)
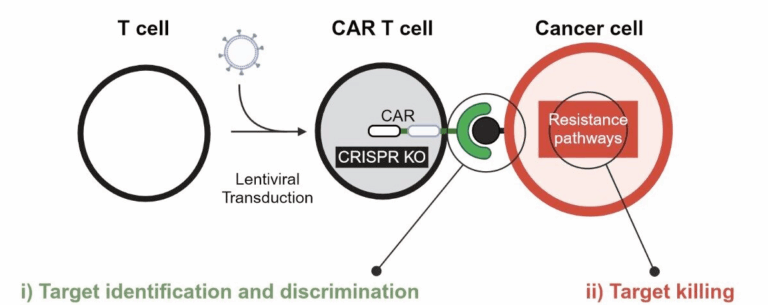
Cellular immunotherapy – especially based on chimeric antigen receptor (CAR) T cells – has revolutionized cancer therapy in recent years. However, we need to overcome many challenges to harness the full potential of therapeutic T cells, specifically in myeloid malignancies and solid tumors. In our newly established lab, we are working on understanding and manipulating the mechanisms of CAR T-cell killing with the ultimate goal of contributing to the next generation of cellular therapies for our patients.
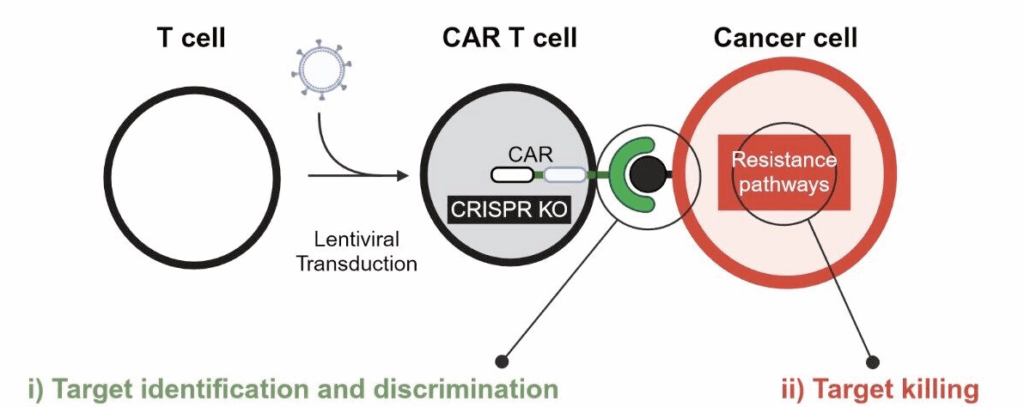
Our group is working on two main projects:
- i) CAR T cells need to discriminate between healthy and malignant tissue to eliminate cancer cells from the body without causing harmful side effects. We are exploring a novel anti-cancer target in the CAR T-cell format.
- ii) Once the CAR T cell identifies cancerous target cells, it has to perform its cytotoxic function, overcome potential resistance mechanisms and kill tumor cells. The second goal of our lab is to understand and manipulate this T cell-target cell dynamic.
We are addressing these challenges by developing and using in vitro and in vivo tools in collaboration with international colleagues at the Institute of Molecular Biochemistry at the Biocenter Innsbruck and at the Department of Hematology and Oncology at the University Hospital Innsbruck. We have close and fruitful collaborations with clinicians and basic researchers of the MUI. This synergy of scientific and clinical expertise is putting us in the optimal position to undertake our highly ambitious projects.
Pictures
Selected Publications
The Dsc ubiquitin ligase complex identifies transmembrane degrons to degrade orphaned proteins at the Golgi. Weyer Y, Schwabl SI, Tang X, Purwar A, Siegmann K, Ruepp A, Dunzendorfer-Matt T, Widerin MA, Niedrist V, Mutsters NJM, Tettamanti MG, Weys S, Sarg B, Kremser L, Liedl KR, Schmidt O, Teis D. Nat Commun. 2024 Oct 26;15(1):9257. doi: 10.1038/s41467-024-53676-6. PMID: 39461958 Free PMC article.
TXNIP mediates LAT1/SLC7A5 endocytosis to reduce amino acid uptake in cells entering quiescence. Kahlhofer J, Marchet N, Seifert B, Zubak K, Hotze M, Egger A, Manzl C, Weyer Y, Weys S, Offterdinger M, Herzog S, Reiterer V, Kwiatkowski M, Wortmann SB, Nemati S, Mayr JA, Zschocke J, Radlinger B, Thedieck K, Huber LA, Farhan H, de Araujo ME, Kaser S, Scholl-Bürgi S, Karall D, Teis D. bioRxiv; 2024. doi: 10.1101/2024.10.29.620655. PPR:PPR933449.
The structure of the Orm2-containing serine palmitoyltransferase complex reveals distinct inhibitory potentials of yeast Orm proteins. Körner C, Schäfer JH, Esch BM, Parey K, Walter S, Teis D, Januliene D, Schmidt O, Moeller A, Fröhlich F. Cell Rep. 2024 Aug 27;43(8):114627. doi: 10.1016/j.celrep.2024.114627. Epub 2024 Aug 20. PMID: 39167489 Free article.
Functional characterization of TMEM86A and TMEM86B mutants by a novel lysoplasmalogenase assay. Kummer D, Dorigatti I, Dunzendorfer-Matt T, Golderer G, Werner ER, Watschinger K. J Lipid Res. 2025 Feb 28:100766. doi: 10.1016/j.jlr.2025.100766. Online ahead of print. PMID: 40024572
Targeting the mevalonate or Wnt pathways to overcome CAR T-cell resistance in TP53-mutant AML cells. Mueller J, Schimmer RR, Koch C, Schneiter F, Fullin J, Lysenko V, Pellegrino C, Klemm N, Russkamp N, Myburgh R, Volta L, Theocharides AP, Kurppa KJ, Ebert BL, Schroeder T, Manz MG, Boettcher S. EMBO Mol Med. 2024 Mar;16(3):445-474. doi: 10.1038/s44321-024-00024-2. Epub 2024 Feb 14. PMID: 38355749
Selection of Funding
FWF 10.55776/P35874: Control of amino acid transporter endocytosis & degradation
FWF 10.55776/P34907: Organelle specific rules for the self-assembly of ESCRT-III
FWF 10.55776/DOC82: Cellular Basis of Diseases: Molecular Control of Metabolism and Inflammation (Coordination)
FWF 10.55776/FG20: Organelle proteostasis in cellular quiescence and growth
FWF 10.55776/P34723: Role of plasmalogens in ether lipid-associated pathologies
FWF 10.55776/FG15: Damage & repair of membrane lipids in health and diseases
FWF 10.55776/I5406: NECESSITY-New chemical entities modulating SARSCoV2 activity
Tiroler Nachwuchsforscher*innenförderung: Unravelling the role of ether lipids in osteogenesis: a multiomics approach
Collaborations
Robbie Loewith, University of Geneva, Geneva, CH
Snezhana Oliferenko, Francis Crick Institute, King’s College, UK
Benoit Kornmann, University of Oxford, Oxford, UK
Tomas Kirchhausen, Harvard Medical School, Cambridge, USA
Florian Fröhlich, Osnabrück University, Osnabrück, DE
Florian Wilfling, MPI Biophysik Frankfurt, DE